New microscope collects dynamic images of the molecules that animate life
Over the last decade, powerful new microscopes have dramatically sharpened biologists' focus on the molecules that animate and propel life. Now, a new imaging platform developed by Eric Betzig and colleagues at the Howard Hughes Medical Institute's Janelia Research Campus offers another leap forward for light microscopy. The new technology collects high-resolution images rapidly and minimizes damage to cells, meaning it can image the three-dimensional activity of molecules, cells, and embryos in fine detail over longer periods than was previously possible.
The developers of the new lattice light sheet microscope have teamed with cell and molecular biologists to produce stunning videos of biological processes across a range of sizes and time scales, from the movements of individual proteins to the development of entire animal embryos. The scientists, including postdoctoral researchers Bi-Chang Chen (now at the Research Center for Applied Sciences in Taiwan), Wesley Legant, and Kai Wang, described their new technology and its applications in Science.
Betzig, one of three scientists sharing the 2014 Nobel Prize in Chemistry, has developed a suite of new imaging technologies over the last 10 years. The techniques have improved biologists' ability to visually track the movements of cells' tiniest structures – but there were always trade-offs. Imaging cells at high resolution in three dimensions usually meant sacrificing imaging speed, as well as subjecting cells to significant light-induced toxicity.
"What happens is you end up designing the questions you ask around the tools that are available," Legant says. With the lattice light sheet, the Betzig team can now optimize their imaging technology for the questions that biologists want to answer.
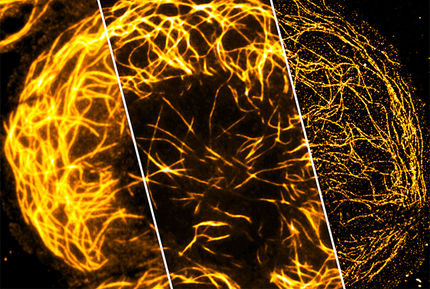
The new microscope evolved from one Betzig unveiled in 2011. The Bessel beam plane illumination microscope illuminates samples with a virtual sheet of light, created when a beam of non-diffracting light called a Bessel beam sweeps across the imaging field. It produces high-resolution images with less light damage than a traditional microscope, and is fast enough to record dynamic processes in living cells.
Betzig's team had to incorporate a few tricks into that microscope to address a problem that arose from the shape of the Bessel beam. "It's not just a thin pencil of light – it has these dimmer side lobes," Betzig explains. "So when you sweep that across a sample, you get out-of-focus light." One solution was to shift the beam step-by-step across the sample, illuminating it with a grating pattern. The researchers could then computationally strip out the blurriness caused by the Bessel beam's side lobes – a technique known as structured illumination.
Structured illumination can also be used to overcome light microscopes' usual limits of spatial resolution. To apply a super-resolution structured illumination technique developed at Janelia by the late Mats Gustafsson, Betzig's team moved the Bessel beam to produce a lattice-like pattern of light. "With that we not only get rid of the side lobe stuff, we actually push the resolution a bit beyond the diffraction limit," he says.
To reduce the time required to move the Bessel beam each time a sample was imaged, the developers split the beam into seven parallel parts, so each traveled just one-seventh of the original distance. Suddenly, the cells they were imaging seemed healthier. "What was shocking to us was that by spreading the energy out across seven beams instead of one, the phototoxicity went way down," Betzig says. "What I learned from that experience is that while the total dose of light you put on the cell is important, what's far more important is the instantaneous power that you put on the cell."
Could they reduce light toxicity even more by dividing the Bessel beam? Betzig wasn't sure what would happen when the beams began to crowd one another, so he modeled the potential effects. Although the beams interfered with one another, the model proposed certain arrangements where that interference actually destroyed the undesirable lobes of light. If he could reproduce those "magic periods" with actual light, the imaging potential was huge. "What you get is sort of a triple win," he says. "You spread the energy out, this highly structured plane is ideal for doing structured illumination with high contrast, and you get rid of that nasty side lobe problem."
That's when Betzig dusted off an idea he'd had more than 10 years ago. Working theoretically, he'd proposed a technique he called optical lattice microscopy, which would limit damage to cells by illuminating a sample with a massive three-dimensional array of light foci. Other technologies had taken hold when Betzig moved to Janelia in 2005, and he'd never built an optical lattice microscope. But the idea remained sound. "We used the lattice theory to predict the patterns that would produce these magic periods," Betzig says.
Moving from theory to actual patterns of light proved challenging. Chen and Wang tackled the problem, and eventually devised a way to create the optical lattice by modulating light directly on the imaging plane. After months of struggle, Chen says, they were able to produce three-dimensional images of cells with the detail, speed, and low toxicity that they'd hoped for. "When we saw the first experimental data, we knew everything was working fine," Chen says.
The new microscope operates in two modes. One uses the principles of structured illumination to create very high-resolution images. In this case, the final image is created by collecting and processing multiple images of every plane of the sample. Imaging can be sped up to capture faster processes, albeit at lower resolution, with an alternative "dithered" mode. Light exposure, and thus damage to cells, is lower in the dithered mode; in many cases, tagged proteins are naturally replaced by cells before their signal fades appreciably. "So there are many cells you could look at forever in 3D," Betzig says.
The microscope's high resolution means it can track the movements of individual proteins in three dimensions, and Betzig says these applications are among the most exciting. "Normally when people do single-molecule studies, they have to do them in thin, flat cells, because the out-of-focus light kills you if it's thick." The lattice's ultra-thin light sheet eliminates that problem, and permits single molecules to be seen even in large multicellular specimens. The microscope is also fast enough to track the rapid growth and retraction of cytoskeletal components in dividing cells, and gentle enough to monitor the molecular dynamics of developmental processes that unfold over many hours.