Machinery of epigenetic inheritance
Relevant to development and cancer
Scientists at Indiana University have unlocked one of the mysteries of modern genetics: how acquired traits can be passed between generations in a process called epigenetic inheritance. The new work finds that cells don't know to silence some genes based on information hardwired into their DNA sequences, but recognize heritable chemical marks that are added to the genes. These chemical tags serve as a form of molecular memory, allowing cells to recognize the genes and remember to silence them again in each new generation.
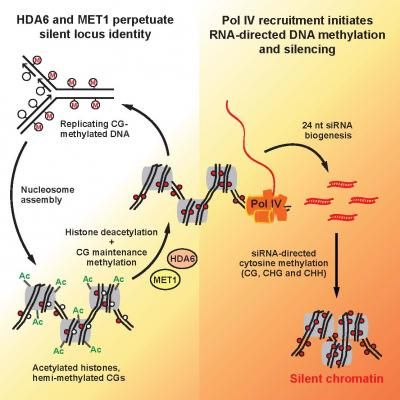
Epigenetic inheritance is a two-step process, with a heritable molecular memory first forming to maintain a chromatin state required later for actual silencing of a genetic locus.
Craig Pikaard and Todd Blevins
The discovery made by a 12-member all-Indiana University team of scientists led by IU biologist and biochemist Craig Pikaard provides important new insight into how plant cells know to silence a genetic locus -- that specific place on a chromosome where a gene is located -- in every successive generation.
Rather than rely on intrinsic, DNA sequence-based information, the cells instead must recall the need to silence specific loci by relying on chemical marks displayed on the complex of DNA and proteins called chromatin. Addition, or removal, of one-carbon (methyl) or two-carbon (acetyl) chemical tags are ways of modifying chromatin that can impart additional, epigenetic (literally, "above genetic") information to a locus beyond the genetic information encoded in the DNA.
The ability to perpetuate chromatin marks serves as a form of epigenetic memory that confers what Pikaard calls silent locus identity, a pre-established state that is needed for the cell to deliver to the loci the machinery that actually accomplishes silencing in a multi-step process known as RNA-directed DNA methylation (RdDM). RdDM involves short-interfering RNAs (siRNA), tiny RNA molecules that are 24 nucleotides long and that guide the addition of methyl groups to matching DNA strands, ultimately rendering the genes inactive.
"Importantly, this work shows that silent locus identity is required for, but separable from, actual gene silencing," Pikaard said. "We've found that epigenetic inheritance is a two-step process, with the heritable specification of silent locus identity occurring before actual silencing of the locus can occur."
Scientists are interested in epigenetic inheritance because it's a process by which heritable modifications occur in gene function without changes in the base sequence of an organism's DNA being required. Disease states such as cancer, which occur sporadically during an individual's lifetime, are increasingly recognized as having an epigenetic basis.
Pikaard said the new work not only sheds important new light on the mechanisms responsible for epigenetic inheritance, a topic of broad interest in the fields of genetics and chromosome biology, but it also helps explain the basis for the recruitment of two plant-specific gene silencing enzymes -- the RNA polymerases Pol IV and Pol V -- first identified by Pikaard in 1999.
Specifically, the researchers tested and identified the relationship between histone deacetylase 6 (HDA6), an enzyme that removes acetyl groups from histones, and the CG DNA sequence maintenance methyltransferase, MET1, and discovered that their partnership in maintenance methylation can explain the perpetuation of epigenetic memory that accounts for silent locus identity.
"Collectively, our results show that silent locus identity is perpetuated from generation to generation through the actions of HDA6 and MET1," Pikaard said. "These activities are not sufficient to silence the loci but maintain a chromatin state that is required for Pol IV recruitment, siRNA biogenesis and RdDM, which is what ultimately silences the loci."
When the team removed the RdDM pathway in Pol IV and Pol V mutant strains of the model plant Arabidopsis thaliana (rockcress), all gene silencing was lost, but silent locus identity remained. They then removed the HDA6 and MET1-dependent process that specifies silent locus identity and, importantly, the epigenetic memory required for silent locus identity was lost and unable to be regained.