A study by IRB Barcelona sheds light on the reasons behind genomes differences between species
All living organisms on Earth are divided into three large domains: archaea, bacteria and Eukarya, and from the beginning of life – more than 3,000 million years ago -, the genomes of each group have evolved towards distinct structures that have favored their separation. A study led by Lluís Ribas de Pouplana, ICREA researcher at the Institute for Research in Biomedicine (IRB Barcelona) and head of the Gene Translation Laboratory, gives an explanation for the divergent evolution of the genomes of different groups of species. The connection between the function of enzymes and the composition of the genomes shed light on the evolution and structure of genes, and explains differences between archaebacteria, bacteria and eukaryotes.
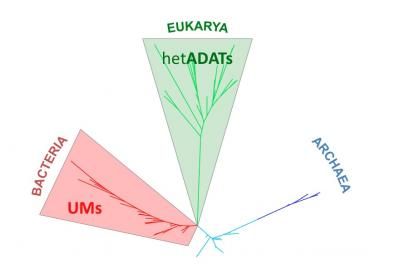
This is a phylogenetic tree based on genome tRNA composition separates Archaea, Bacteria and Eukarya species. The tree shows that the appearance of several enzymes that modify tRNAs in each domain, UMs in Bacteria and hetADATs in Eukarya, have exerted evolutionary pressure, which has contributed to the separation of species.
Eva Novoa (IRB Barcelona)
The scientists have analyzed the distribution and abundance of transfer RNA genes – tRNA - in more than 500 species belonging to the three domains of life. They discovered that the structure of genomes was adapted to the activity of some enzymes, which differ for Bacteria and for Eukarya and are absent in Archaea.
The activity of these enzymes modifies tRNAs, allowing them to recognize up to three distinct codons. The activity of the bacterial and eukaryal enzymes is different, which explains why the genomes and the gene composition of bacteria, eukarya, and archaea have diverged. This discovery furthers our understanding of the relation between genome structure and the speed of protein synthesis from its genes. So, for example, as Ribas de Pouplana explains, "genes rich in codons that can be read by modified tRNAs have high expression levels. Or said the other way around, the greater the abundance of a protein in a cell the higher the number of triplets found in its gene sequence that can be read by modified tRNAs. Our findings contribute to the understanding of how the translation machinery works and explains why genomes for each group of species have a distinct codon composition."
This finding paves the way to many applications. One of these is in biotechnology as the discovery of the relevance of these modifications will allow an improvement in the industrial production of proteins: "We now have another parameter with which to optimize the synthesis of proteins from a gene", explains Eva Novoa, the first author of the article, who started her PhD studies in 2008 in Ribas' lab through the "la Caixa" International PhD Programme in Biomedicine. "To give just one example, human insulin is "manufactured" in bacteria and our discovery would allow this production to be increased if we take into account the activity of these enzymes", says Novoa. The finding is also relevant for the study of cancer: "it is possible that these modification enzymes are over-represented in some kinds of cancer. In fact, this would be logical because cancer cells are highly efficient in producing proteins."
Published in Cell, the article demonstrates how organisms have evolved in a different manner to achieve better adaptations and to have optimum protein translation efficiency. "We don't exactly know why these enzymes appear or why they are different in bacteria and in eukaryotes but it's clear that they contribute to the separation of genomes of these two groups. The genetic code is the same but what has changed is the relative importance of different codons of the code. And this made the genome of these groups differentiate", concludes Ribas.