Scientists X-ray anti-inflammatory drug candidates
Study resolves molecular structures of Spiegelmers for the first time
Using DESY's ultra bright X-ray source PETRA III, scientists have decoded the molecular and three-dimensional structure of two promising drug candidates from the new group of Spiegelmers for the first time. The results provide a deeper understanding of the mode of action of these substances that have already entered clinical trials. The researchers from the Universities of Hamburg and Aarhus (Denmark) together with colleagues from the biotech company NOXXON in Berlin present their work in the journal Nature Communications.
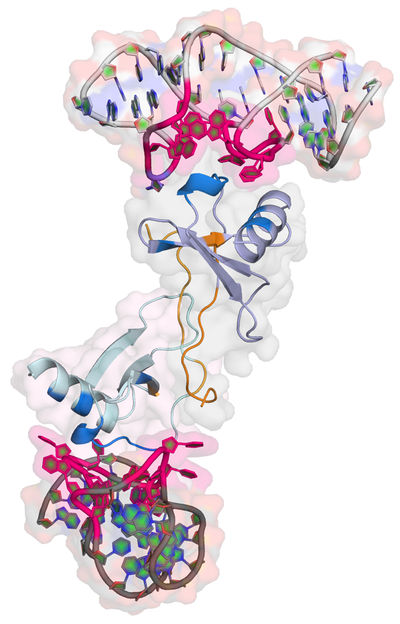
Structure of the Spiegelmer NOX-E36 bound to its target protein CCL2.
Dominik Oberthür/CFEL
Spiegelmers are a young group of promising pharmaceutical substances. They rely on the same building blocks as the nucleic acids RNA and DNA that fulfil various tasks in the organism - from storing genetic information and messaging to the regulation of genes. Artificial RNA or DNA molecules called aptamers can be tailored to bind to certain proteins with high specificity, blocking their function. Aptamers are well tolerated in the organism as they consist of natural building blocks. For these reasons, aptamers are seen as promising drug candidates. Since 2006, an aptamer for the treatment of age-related macular degeneration AMD, an eye condition that can lead to blindness, is approved and on the market.
Usually, RNA and DNA molecules are quickly degraded by enzymes within the body. This severely limits their application as pharmaceutical drugs. However, most biomolecules come in two mirror-image variants, the L-form and the D-form. Natural nucleic acids always exist in the D-form, while proteins are always build in their L-form in the body. Artificial aptamers that are constructed in the naturally not occurring L-form are not degraded by the organism. These mirror-image variants of aptamers are called Spiegelmers. “An advantage of Spiegelmers is that they are not targeted by the body's enzymes,” explains Prof. Christian Betzel from the University of Hamburg.
“Spiegelmers can be identified and optimised in the lab through a sophisticated evolutionary procedure. However, exact structure data of Spiegelmers have not been available until now,” says first author Dr. Dominik Oberthür from the Center for Free-Electron Laser Science CFEL, a cooperation of DESY, Max Planck Society and the University of Hamburg. If the exact structure of a Spiegelmer and its binding site at the target protein is known, its mode of action can be decoded and its structure could be further fine-tuned, if necessary.
The team around Betzel used PETRA III's bright X-rays to analyse the Spiegelmer NOX-E36 from NOXXON. It blocks the protein CCL2 that is involved in many inflammatory processes in the body. “If you target an inflammatory protein with a Spiegelmer, you have a good chance to tone down the inflammation in the body,” notes Betzel. NOX-E36 has already been successfully tested in a phase IIa clinical trial with patients.
In order to analyse the structure of the drug candidate, the scientists first had to grow crystals of the Spiegelmer bound to its target protein CCL2. “Growing these crystals was quite a challenge,” recalls Betzel. Because it contradicts their natural function, most biomolecules are notoriously hard to crystallise.
The crystals were analysed at the PETRA III measuring station P13, run by the European Molecular Biology Laboratory EMBL. Crystals diffract X-ray light, producing a characteristic pattern on the detector. From this diffraction pattern the structure of the crystal's building blocks can be calculated – in this case the Spiegelmer's structure, bound to its target protein. In the same manner, a group around Laure Yatime from the University of Aarhus solved the structure of another Spiegelmer: NOX-D20 binds to the protein C5a that is involved into many inflammatory processes, too. The group also reports the structure in Nature Communications.
The analyses reveal the structure of both Spiegelmers with a spatial resolution of 0.2 nanometres (millionths of a millimetre) – that's on the order of individual atoms. “I am delighted to finally have a high resolution visualization of the remarkable shapes of two Spiegelmer drug candidates,” comments Dr. Sven Klussmann, founder and chief scientific officer of NOXXON, and also co-author on both articles. “The structural data not only provide the first look at the unusual interaction of a mirror-image oligonucleotide with a natural protein but also deepens our understanding of the two molecules’ mode of action.”
Original publication
„Crystal structure of a mirror-image L-RNA aptamer (Spiegelmer) in complex with the natural L-protein target CCL2“; Dominik Oberthür et al.; „Nature Communications“, 2015
„Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer“; Laure Yatime, Christian Maasch, Kai Hoehlig, Sven Klussmann, Gregers R. Andersen & Axel Vater; „Nature Communications“, 2015