Lignin breakthroughs serve as GPS for plant research
Biosystems approach offers step-by-step directions for young scientists
Researchers at North Carolina State University have developed the equivalent of GPS directions for future plant scientists to understand how plants adapt to the environment and to improve plants' productivity and biofuel potential.
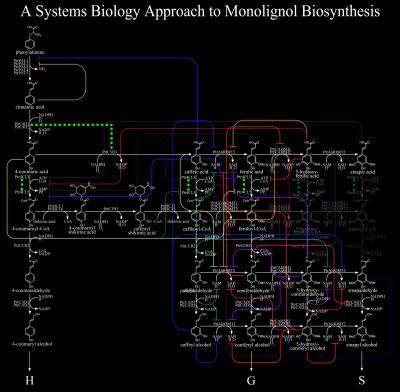
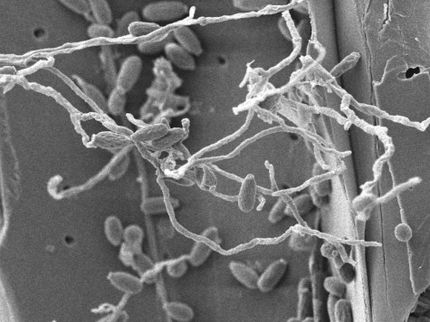
This illustration shows detailed work on lignin biosynthesis from North Carolina State University that offers step-by-step directions for future plant research.
Jack P. Wang
Two articles published in The Plant Cell offer a step-by-step approach for studying plant traits, drawing on comprehensive, quantitative research on lignin formation in black cottonwood. Lignin, an important and complex polymer responsible for plant growth and development, provides mechanical strength and water transport that enables some trees to grow 100 meters tall. However, lignin must be removed for biofuel, pulp and paper production–a process that involves harsh chemicals and expensive treatments.
The interdisciplinary research provides a new approach integrating knowledge of genes, proteins, plant chemical compounds and engineering modeling to understand how plants make products and structures needed for growth and development. This work in the new area of plant systems biology, integrating biology, chemistry and engineering, sets a new standard for understanding any complex biological feature in the future.
"I describe these findings as MapQuest for plant scientists," says Vincent Chiang, co-director of NC State's Forest Biotechnology Group, the lead team for the project, which involved scientists in the College of Natural Resources, College of Engineering and College of Sciences. "For example, the systems biology approach could be applied in research to develop sweeter citrus fruit, disease-resistant rice or drought-resistant trees."
Over many years of intensive research, the interdisciplinary team led by Chiang purified 21 pathway enzymes and analyzed 189 different parameters related to lignin formation. With help from engineering colleagues Cranos Williams and Joel Ducoste, the team developed models that predict how pathway enzymes affect lignin content and composition. One of the enzymes forms a novel four-part structure, which was discovered as part of this work, including quantitation of all 21 enzymes carried out by chemist David Muddiman.
"The model, based on a comprehensive set of equations for each step in the process, can now predict changes in the amount and composition of lignin, as well as why it's often difficult to modify lignin in plants" says Ronald Sederoff, co-director of Forest Biotechnology Group.
The GPS-like findings could reduce years of research time required to make future advances, Chiang says. "We hope that this research will stimulate similar work by young scientists. Don't be discouraged by complex biological processes. Our work shows a successful approach for such studies."
Most read news
Other news from the department science

Get the life science industry in your inbox
From now on, don't miss a thing: Our newsletter for biotechnology, pharma and life sciences brings you up to date every Tuesday and Thursday. The latest industry news, product highlights and innovations - compact and easy to understand in your inbox. Researched by us so you don't have to.