Molecular engines star in new model of DNA repair
Study reveals major role of RNA polymerase and other enzymes in DNA repair
Our health depends in large part upon the ability of specialized enzymes to find and repair the constant barrage of DNA damage brought on by ultraviolet light radiation and other sources. In a new study NYU School of Medicine researchers reveal how an enzyme called RNA polymerase patrols the genome for DNA damage and helps recruit partners to repair it. The result: fewer mutations and consequently less cancer and other kinds of disease.
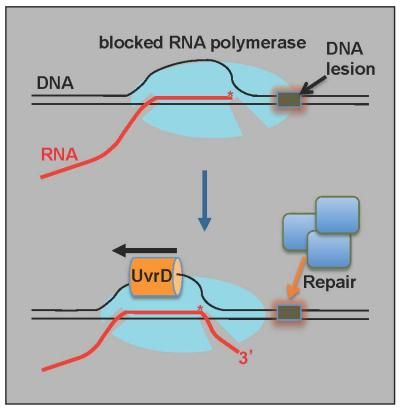
A new model of DNA repair is revealed in a new study. In this model, RNA polymerase patrols tracks of double-stranded DNA and stalls over damaged areas. An enzyme called UvrD helicase pulls the blocked RNA polymerase off the tracks, exposing broken DNA to repair. After the repair, the polymerase continues along the tracks.
NYU Langone Medical Center
The study, led by Evgeny Nudler, PhD, a Howard Hughes Medical Investigator and the Julie Wilson Anderson Professor of Biochemistry at NYU Langone Medical Center, is being published online in the issue of Nature.
Scientists have long known that RNA polymerase slides along the telltale tracks of double-stranded DNA and uses that template to create a growing chain of RNA molecules. This RNA chain, in turn, contains all of the information needed to construct cellular proteins. The enzyme, however, can stall as it patrols the tracks and encounters significant DNA damage. Even worse, it can become lodged over the damaged site, preventing any repair specialists from reaching it.
In the new study, the NYU School of Medicine researchers reveal how another enzyme called UvrD helicase acts like a train engine to pull the RNA polymerase backwards and expose the broken DNA so a repair crew can get to work.
The finding has major implications for a patching mechanism that is widely shared by organisms ranging from bacteria to humans, says Dr. Nudler. "Better repair means fewer mutations, which also means slower aging, less cancer and many other pathologies," he says.
Although the research, conducted in Escherichia coli bacteria, focused on one type of DNA repair, Dr. Nudler says the evidence suggests that other cellular repair pathways might use the same mechanism to recognize and then resolve the damage. Failure to do so can lead to profound consequences: inherited defects in the gene that encodes the human analog of UvrD, a protein known as XPB, have been linked to a range of devastating disorders.
In a condition known as xeroderma pigmentosum, for example, the faulty DNA repair system cannot fix damage caused by ultraviolet radiation. Consequently, any exposure to sunlight can cause serious skin and eye damage and greatly elevate the risk of skin cancer Similarly, children born with Cockayne syndrome age prematurely and are often short in stature due to inadequate DNA repair. Those with a third related condition called trichothiodystrophy have brittle hair, recurrent infections and delayed development.
The study by Dr. Nudler's group and colleagues in Russia used a battery of biochemical and genetic experiments to directly link UvrD to RNA polymerase and to demonstrate that UvrD's pulling activity is essential for DNA repair. The lab results also suggest that UvrD relies on a second factor, called NusA, to help it pull RNA polymerase backwards. Those two partners then recruit a repair crew of other proteins to patch up the exposed DNA tracks before the train-like polymerase continues on its way.
According to Dr. Nudler, his team's study offers a convincing justification for a puzzling phenomenon known as pervasive transcription, which he calls "one of the most enigmatic and debated subjects of molecular biology." The question, he says, boils down to this: Why do RNA polymerases transcribe most of the genome within humans and other organisms, converting vast stretches of DNA to RNA, when only a tiny fraction of those resulting RNA transcripts will ever prove useful? Isn't that RNA polymerase activity a waste of energy and resources?
"Our results imply that a major role of RNA polymerase is to patrol the genome for DNA damage," he says. "This is the only molecular machine that is capable of continuously scanning the chromosomes for virtually any deviation from the canonical four bases in the template strand: A, T, G and C." The polymerase's extensive transcription activity, then, might be well worth the effort if its continuous vigilance also ensures that any DNA damage gets fixed through the assistance of the pulling factors and other collaborators.
In addition to its insights on DNA repair, the paper describes a powerful new method for mapping protein-protein interactions at high resolution. Dr. Nudler says the method, known as chemical cross-linking coupled with mass-spectrometry, or XLMS, can be widely used by other labs and applied to virtually any protein interactions.