Fitting pieces for biosensors
Research and industry are increasingly exploiting the potential of aptamers. As well as their application in research, medical diagnosis and treatment, aptamers are also interesting as a basis for biosensors for use in environmental analysis because their characteristics enable them to identify and bind target molecules as surely as a key fits a lock. In a new book, researchers at the Helmholtz Centre for Environmental Research (UFZ) describe the methods used to obtain aptamers. A newly-approved project aims to develop new nanostructured biosensors to measure harmful substances in water.
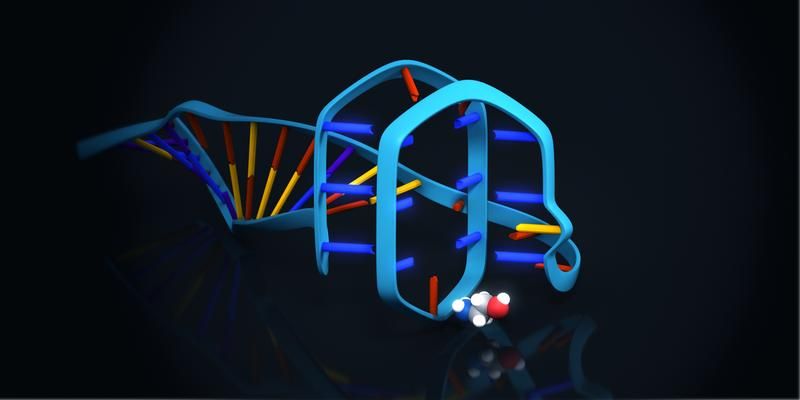
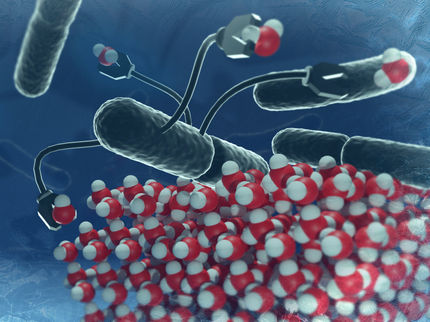
Graphical model of the ethanolamine binding aptamer. The small ethanolamine molecule will be bound to a G-rich sequence region of the aptamer, which is able to form a quadruplex structure. Exemplary illustration of one possible option of binding between aptamer and target molecule.
Ronny Jesse/www.jesse3d.de
The term aptamer means something like "fitting pieces" (from the Latin word aptus, meaning to fit, and the Greek word meros, meaning piece). Aptamers consist of nucleic acids and have a three-dimensional structure that enables them to identify and bind certain target molecules. These binding abilities allow, for instance, tracing, detecting and measuring certain substances. For this purpose aptamers can be used in e.g. biosensors. Biosensors are a simple, quick, low-cost way of taking measurements. At the heart of all biosensors there is a biologically active component. This bioreceptor has the ability to interact with its target substance, producing a signal in the process. Signal transducers in the sensor make the signal measurable and visible. Once measurement has taken place, the biosensor is returned to its original state. In other words, it can be regenerated.
To design biosensors, scientists need suitable bioreceptors that can identify the target substance. Aptamers offer great potential for use as biological recognition elements in biosensors. Firstly, however, the scientists have to identify the right aptamer for a particular target molecule. Such target molecules can be very complex structures, like whole cells or organisms, or tiny molecules consisting of just a few atoms. The search for the right aptamers is like managing a gigantic molecular dating agency. Using an in vitro method, researchers select the best binding partners for the target molecule from a huge pool of 10 million x 100 million nucleic acids with different sequences. This selection method is called SELEX (Systematic Evolution of Ligands by EXponential enrichment). SELEX is an evolutionary process performed in a 'test tube' (in vitro). Therefore the use of animals, plants or cell cultures is not required during the process. Beside ethical benefits, another advantage of this is the possibility of aptamer selection even for toxic substances. Once suitable binders (aptamers) for a target molecule have been found by the SELEX process and their sequences have been defined, aptamers can be produced very accurately and reliably at any time by chemical synthesis. Accordingly no biologically induced variations have to be taken into account during synthesis, as they would if natural systems were used. Another advantage is the possibility to give the aptamers defined properties relatively easily by sequence modifications or by adding functional groups or reporter molecules. The added properties can enable or simplify the measuring process, or improve the stability of the aptamers.
In the UFZ's biosensor laboratory headed by Dr Beate Strehlitz, Dr Regina Stoltenburg has developed two different modifications of the SELEX method. One of these is known as FluMag SELEX. The 'Flu' stands for fluorescence and refers to the fact that a fluorescence molecule is added to the nucleic acids during the SELEX procedure to make them visible. In this manner the molecules can always be found again and researchers can measure the enrichment of those which exhibit best binding and detecting abilities to the given target. The 'Mag' refers to magnetic beads. These are dust-mote-sized magnetic beads onto which the scientists 'stick' the even smaller target molecules to make them more manageable.
Many other modifications of the SELEX procedure have been developed by teams in research institutes around the world enabling aptamer selections for a wide range of different applications. In the new book "Aptamers in Bioanalysis" Beate Strehlitz and Regina Stoltenburg describe the SELEX procedure and its many variants in a review chapter.
By use of the FluMag SELEX procedure, developed by the UFZ research group in Leipzig, aptamers for a wide range of target molecules can be selected. Thus it has been used successfully to generate aptamers for a protein, a peptide and for ethanolamine, the smallest molecular aptamer target so far. The ethanolamine-binding aptamers have been patented. Additionally Dr Christine Reinemann succeeded in selecting aptamers for soluble constituents of Penicillium expansum spores (mould spore extract). Based on this aptamers she hopes to develop a detection method for mould fungi within a project with third-party funding from Saxony's Office for Environment, Agriculture and Geology (LfULG). Mould fungi are one of the reasons behind the increase in allergies in Germany. Together with PhD student Sören Linkorn and researchers from the Institute of Food Technology and Bioprocess Engineering at TU Dresden, Dr Regina Stoltenburg wants to select aptamers that can recognise pathogenic bacteria. These aptamers are supposed to develop a biosensor-based detection method for pathogens in water. This research is being conducted within the International Water Research Alliance Saxony (IWAS). A quick method for identifying these harmful germs is particularly important in arid parts of the world because contaminated drinking water in these areas can cause diseases, death and even epidemics.
The German Federal Ministry of Education and Research (BMBF) has just approved a joint project under leadership of Forschungszentrum Dresden-Rossendorf (FZD) and in collaboration with the University of Rostock, proaqua GmbH & Co. KG in Mainz and the UFZ. The project is part of the BIONA (Bionic Innovations for Sustainable Products and Technologies) research programme. It will use the natural nanostructures of bacterial coat proteins to fix aptamers onto sensor surfaces in a controlled manner. The UFZ is to develop aptamers that are capable of detecting certain organic substances, such as undesirable pharmaceutical residues, that enter the environment through wastewater.
If the UFZ researchers are successful, in a few years' time, biosensors will be able to help with the prompt identification of potential health risks, such as mould fungi in rooms or germs and pharmaceutical residues in water.
Original publication: Strehlitz, B., Stoltenburg, R.; "SELEX and its Recent Optimizations" in: Mascini, M.; "Aptamers in Bioanalysis"; WILEY Interscience 2009