“Two in One” enzyme: unusually flexible Scientists from the RUB have solved the structure of a viral protein
The Journal of Biological Chemistry has ranked this documentation as “Paper of the Week.”
A virus that infects the marine cyanobacterium Prochlorococcus can produce specific pigments more effectively than its host can. It requires only one enzyme, in contrast to the host Prochlorococcus, which needs two enzymes. The virus makes use of phycoerythrobilin synthase, a “two in one” enzyme. Within the frameworks of his dissertation, Thorben Dammeyer, a member of the research team under the supervision of Prof. Nicole Frankenberg-Dinkel (Physiology of Microorganisms) and Assistant Professor Dr. Eckhard Hofmann (X-ray diffraction analysis of proteins), solved the 3D structure of the enzyme. An unexpected flexibility was discovered, allowing sections of the protein to assume different positions – an unusual property for proteins in combination with their substrate. The scientists have documented their results, honored as “Paper of the Week,” in the current issue of the Journal of Biological chemistry.
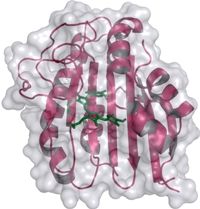
Strukturmodell der Phycoerythrobilin Synthase - 3D structural model of the phycoerythrobilin synthase (PebS) with the attached biliverdin substrate (green).
Prof. Nicole Frankenberg-Dinkel, Faculty for Biology und Biotechnology, Physiology of Microorganisms, Ruhr-University Bochum
Pigments are produced in two steps
The so-called P-SSM2 virus with the “two in one” enzyme infects the cyanobacterium Prochlorococcus, a cyanobacterium found in extremely large numbers in the worlds oceans. The virus does however differ in that - in contrast to its cyanobacterial relatives - it does not harvest light for photosynthesis via red and blue pigments, but with chlorophyll, as is the case with higher plants. Nevertheless Prochlorococcus contains all the genetic information for the entire machinery required to produce these pigments. This takes place in two steps with two different enzymes as catalysts.
Green turns red in one step
Nicole Frankenberg-Dinkel stated that “we have discovered the genetic blueprint for an enzyme within the virus. This enzyme is capable of producing the red pigment more effectively than its host, which has convinced us that the pigment cannot be unimportant for Prochlorococcus, even if it is not required for light trapping. On the other hand, we obviously wanted to know how this enzyme can combine two functions.” The scientists used X-ray diffraction analysis to determine the 3D structure of the enzyme at atomic resolution both alone and in complex with its natural substrate, the green biliverdin IXα. This molecule was found in the binding pocket of the protein, where the conversion into a red pigment takes place. Prof. Frankenberg-Dinkel explained that the scientists were able to observe how different parts of the enzyme around the binding pocket are capable of assuming different positions. “This property might not be unusual for proteins in solution, but is extremely rarely found in protein crystals.” The structural variations observed supplied the scientists with the first indications of the movements of the enzyme during catalysis.
Next step: tracking the evolution
The next stage of research will consist of studies of targeted and randomly genetically altered forms of the unusually flexible protein. Using this system, the scientists want to observe the in vitro evolution of this specific enzyme. Nicole Frankenberg-Dinkel’s and Eckhard Hofmann’s research teams are funded by the Collaborative Research Centre 480 “Molecular Biology of Complex Functions in Botanical Systems.”
Original publication: Dammeyer, T., Hofmann, E. & Frankenberg-Dinkel, N. (2008): Phycoerythrobilin synthase (PebS) of a marine virus. “Crystal structure of the biliverdin-complex and the substrate free form.” J.Biol. Chem. 283, 27547-27554.




















































