Uncovering bacterial cell wall secrets to combat antibiotic resistance
Cell walls -- the jacket-like structures that surround all known bacteria -- may turn out to be bacteria's undoing, holding the key to developing new drugs that target it for destruction.

In the battle against drug-resistant bacteria, Marcos Pires studies the chemical biology of bacterial cell surfaces to better understand how they function -- and possibly how to manipulate them
Illustration by Hvass & Hannibal courtesy of Lehigh University
That perspective is shared by many in the medical and scientific communities, including Marcos Pires . Pires, a biochemist at Lehigh University , is spearheading a novel approach to understanding bacterial cell wall changes in response to antibiotics that could be critical to new drug design--an urgent need in light of the growing threat of antibiotic resistance. His approach is so promising it has recently been recognized by the National Institutes of Health with a Maximizing Investigators' Research Award (MIRA).
Antibiotic resistance occurs when bacterial cells adapt to evade a drug designed to kill it. Making changes to the cell wall is one way bacteria accomplish this. Little is known, however, about just how these structures respond when under attack.
With the 5-year $1.94 million MIRA grant, Pires's group will delve deeply into this process through a unique approach that essentially tricks bacteria into revealing where its cell wall is most vulnerable. Such knowledge could help scientists design next-generation antibiotics that circumvent drug resistance mechanisms.
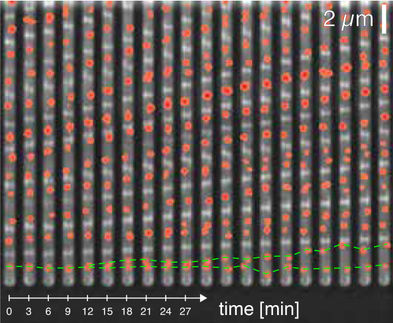
The centerpiece of the research is a process that Pires and his team conduct facilitating live bacteria's absorption of synthetic cell wall fragments constructed in the lab. These fragments are modified with reporter units which then allow researchers to observe, in live bacteria, components of the cell wall machinery under various conditions.
"Bacterial cell walls are unique in their structure and function and are essential to bacterial cells -- making them unique targets for the development of antibiotics," said Pires, assistant professor in the Department of Chemistry. "By 'tricking' bacteria into using some of our cell wall building blocks, we get an unprecedented perspective on how they change when challenged with antibiotics."
MIRA is a program of the National Institute of General Medical Science (NIGMS), a division of NIH that provides support for basic research that increases understanding of biological processes and lays the foundation for advances in disease diagnosis, treatment and prevention. According NIGMS, the goal of MIRA is to increase the efficiency of NIGMS funding by providing investigators with greater stability and flexibility, thereby enhancing scientific productivity and the chances for important breakthroughs.
Identifying bacterial cell wall changes that cause antibiotic resistance
The stakes for drug design breakthroughs to treat drug-resistant bacteria are high. Every year in the United States, more than 2 million people are afflicted with resistant bacterial infections. An estimated 23,000 American lives--and 700,000 lives worldwide--are lost yearly as a result of bacterial infections resistant to current antibiotic treatments. These numbers are only expected to grow.
Bacterial cell walls are the target of some of the most powerful antibiotics discovered to date. Cell wall-targeting antibiotics include some commonly prescribed treatments such as penicillin and amoxicillin. Drugs that target bacteria's cell walls are also among the safest as human cells do not have cell walls and are thus unaffected by the treatment.
According to Pires, individual components of the bacterial cell wall machinery are key to bacteria's adaptation response and, therefore, to drug-resistance. One of his team's goals is to identify the cell wall components that bacteria need to successfully adapt and evade the drugs designed to destroy it.
"If we can identify these 'weak spots', said Pires, "we should be able to find ways to inactivate or circumvent them."