A protein-folding chaperone's functional dance simulated
Using a combination of computational and experimental techniques, a research team at the University of Massachusetts Amherst led by molecular biologist Lila Gierasch has demystified the pathway of interdomain communication in a family of proteins known as Hsp70s - a top target of dozens of research laboratories trying to develop new anti-cancer drugs, antibiotics and treatments for Alzheimer's and Parkinson's diseases.
Gierasch says, "This one presented us with quite a scientific challenge, because all the techniques we usually use to look at communication between the different domains in proteins get stuck when the targets are flexible and not rigid, and the interdomain linker in Hsp70s is very flexible. We had to be clever in our approach."
As she explains, heat shock proteins in the Hsp70 family - molecular weight 70 - are "a really important class of molecular chaperones that have many important jobs in the cell, including binding to client proteins to assist their folding, or to keep them from pathologically aggregating, or to keep them unfolded so they can pass threadlike through a membrane."
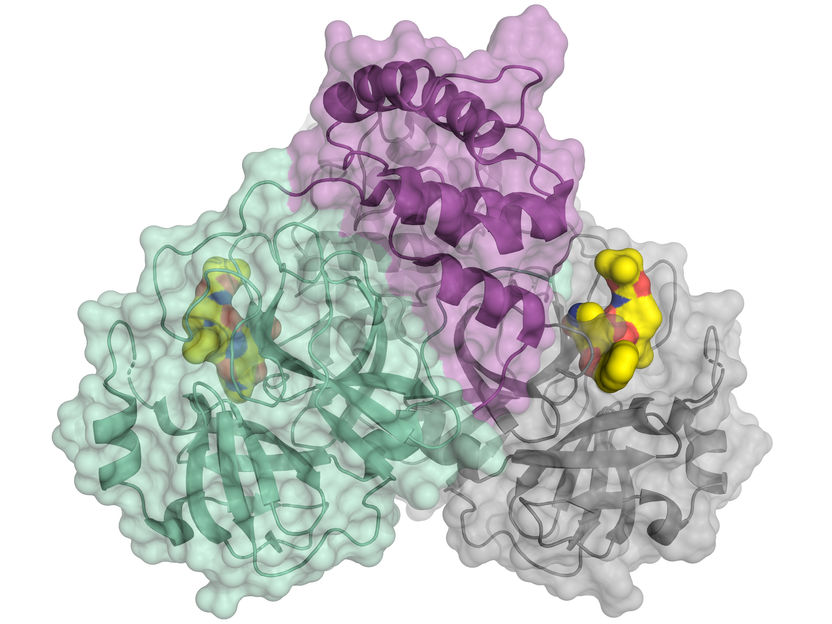
She describes the three parts of a folded Hsp70 protein as a nucleotide-binding domain and substrate-binding domain linked by a "mysterious" interdomain linker, which becomes part of the structure when a small molecule, adenosine triphosphate (ATP), binds to the Hsp70. When ATP releases energy for the cell's use, it changes to adenosine diphosphate (ADP). "You can think of this molecular machine like a Rube Goldberg apparatus," she notes. "We've known for years that the little linker that connects the two domains is important, and when ATP is present it is a part of the two-domain collapsed structure. But with ADP present, the domains move independently. The linker's role in this case was unknown, and posed a puzzle that was very hard to figure out."
Faced with limited direct experimental methods to watch the linker's movements, Gierasch and postdoctoral researcher Charles English, collaborating with adjunct faculty member Woody Sherman, also CSO of Silicon Therapeutics, Boston, conducted a computational study to understand the range of possible linker positions. They used a technique called molecular dynamics, which uses Newton's equations of motion to simulate how atoms and molecules move. This was aided by the massive computing power of graphics processing units and the nearby multi-university Massachusetts Green High Performance Computing Center in Holyoke.
Gierasch says Wenli Meng, a master of nuclear magnetic resonance (NMR) techniques, obtained NMR data that validated the simulations. The linker is just 12 amino acids long, and Meng was able to directly extract linker information from among 636 amino acids. "That was a significant contribution to our work," she notes. "It gave us an experimental fingerprint to compare with what the computer simulation said, and we were delighted by the tight agreement. The combination of NMR data with our simulations made our findings much stronger. Integration of multiple disciplines is greatly advancing life sciences research like this."
She reports, "It turns out that when you look at this linker carefully, it's like a universal joint in a car." For Gierasch, who rebuilt an MG from parts as a graduate student, this analogy is not as unlikely as one might think. "In the driveshaft mechanism, there are rigid pieces connected by joints, and that's what this linker has, three rigid pieces connected by flexible hinges. The result is that the two domains tethered by the linker have restricted rotation with respect to each other. So this protein linker is not like a noodle or piece of yarn. It is rigid in some places yet flexible in others, and, to facilitate proper function of the chaperone, it can twist in ways that facilitate the chaperone's mechanism, but it can't go everywhere."
An "unanticipated bonus" she adds, is discovering that the linker made frequent pauses at a pocket on the substrate-binding domain, "that gives you a potential binding site to target with a drug. We didn't know about this little pocket before, but now we can imagine a way to stop the chaperone-assisted folding cycle by designing a little molecule that will bind at that site."
Gierasch acknowledges that this advance is, by itself, just one step in a long process, in fact one she has been working out for decades. "But that's how science works, one step at a time. With each piece of the puzzle you put in, it clarifies the larger picture." She and co-authors write that enhanced understanding of the interdomain linker's role they have now characterized "fills one more gap in comprehending how these molecular machines perform their myriad physiological functions." This work should be useful for research on Hsp70 targets for cancer, Alzheimer's and Parkinson's disease and developing new antibiotics, they add.
"In addition, seemingly flexible linkers are widespread in signaling molecules in biology, and our work illustrates the importance of studying the roles of these ostensibly flexible units in depth. Delineating their conformational landscapes will shed light on how they may relay information from one region of a molecule to another."
Gierasch adds, "It's said that cancer cells are absolutely addicted to Hsp70s because they are making many proteins in large quantity, and these chaperones are greeting every newly synthesized protein and facilitating their adoption of a proper structure, which is why people are so interested in them. If you inhibit them you would slow the growth of cancers. The problem is that you also would make some other cells sick." Also, new antibiotics might be developed from inhibiting specific bacterial Hsp70s, and because Hsp70s help to protect cells against protein aggregation, inhibiting or manipulating them might lead to new treatments for the rare diseases resulting from protein clumping.