New tool to distinguish between viral, bacterial infections
antibiotics are lifesaving drugs, but overuse is leading to one of the world's most pressing health threats: antibiotic resistance. Researchers at the University of Rochester Medical Center are developing a tool to help physicians prescribe antibiotics to patients who really need them, and avoid giving them to individuals who don't.
Scientists from the University's National Institutes of Health-funded Respiratory Pathogens Research Center identified 11 genetic markers in blood that accurately distinguished between viral and bacterial infections (antibiotics help us fight bacterial infections, but aren't effective and shouldn't be used to treat viruses). The finding is important because physicians don't have a good way to confirm bacterial infections like pneumonia and more-often-than-not default to an antibiotic.
"It's extremely difficult to interpret what's causing a respiratory tract infection, especially in very ill patients who come to the hospital with a high fever, cough, shortness of breath and other concerning symptoms," said Ann R. Falsey, M.D., lead study author, professor and interim chief of the Infectious Diseases Division at UR Medicine's Strong Memorial Hospital. "My goal is to develop a tool that physicians can use to rule out a bacterial infection with enough certainty that they are comfortable, and their patients are comfortable, foregoing an antibiotic."
Falsey's project caught the attention of the federal government; she's one of 10 semifinalists in the Antimicrobial Resistance Diagnostic Challenge, a competition sponsored by NIH and the Biomedical Advanced Research and Development Authority to help combat the development and spread of drug resistant bacteria. Selected from among 74 submissions, Falsey received $50,000 to continue her research and develop a prototype diagnostic test, such as a blood test, using the genetic markers her team identified.
A group of 94 adults hospitalized with lower respiratory tract infections were recruited to participate in Falsey's study. The team gathered clinical data, took blood from each patient, and conducted a battery of microbiologic tests to determine which individuals had a bacterial infection (41 patients) and which had a non-bacterial or viral infection (53 patients). Thomas J. Mariani, Ph.D., professor of Pediatrics and Biomedical Genetics at URMC, used complex genetic and statistical analysis to pinpoint markers in the blood that correctly classified the patients with bacterial infections 80 to 90 percent of the time.
"Our genes react differently to a virus than they do to bacteria," said Mariani, a member of the Respiratory Pathogens Research Center (RPRC). "Rather than trying to detect the specific organism that's making an individual sick, we're using genetic data to help us determine what's affecting the patient and when an antibiotic is appropriate or not."
Falsey, co-director of the RPRC, and Mariani say that the main limitation of their study is the small sample size and that the genetic classifiers selected from the study population may not prove to be universal to all patients.
A patent application has been filed for their method of diagnosing bacterial infection. Edward Walsh, M.D., professor of Infectious Diseases, and Derick Peterson, Ph.D., professor of Biostatics and Computational Biology at URMC, also contributed to the research.
According to the Centers for Disease Control and Prevention, antibiotic resistant bacteria cause at least 2 million infections and 23,000 deaths each year in the United States. The use of antibiotics is the single most important factor leading to antibiotic resistance around the world.
Original publication
Soumyaroop Bhattacharya, Alex F. Rosenberg, Derick R. Peterson, Katherine Grzesik, Andrea M. Baran, John M. Ashton, Steven R. Gill, Anthony M. Corbett, Jeanne Holden-Wiltse, David J. Topham, Edward E. Walsh, Thomas J. Mariani & Ann R. Falsey; "Transcriptomic Biomarkers to Discriminate Bacterial from Nonbacterial Infection in Adults Hospitalized with Respiratory Illness"; Scientific Reports; 2017
Most read news
Original publication
Soumyaroop Bhattacharya, Alex F. Rosenberg, Derick R. Peterson, Katherine Grzesik, Andrea M. Baran, John M. Ashton, Steven R. Gill, Anthony M. Corbett, Jeanne Holden-Wiltse, David J. Topham, Edward E. Walsh, Thomas J. Mariani & Ann R. Falsey; "Transcriptomic Biomarkers to Discriminate Bacterial from Nonbacterial Infection in Adults Hospitalized with Respiratory Illness"; Scientific Reports; 2017
Topics
Organizations
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.
Most read news
More news from our other portals
Last viewed contents
Marshall-Smith_syndrome
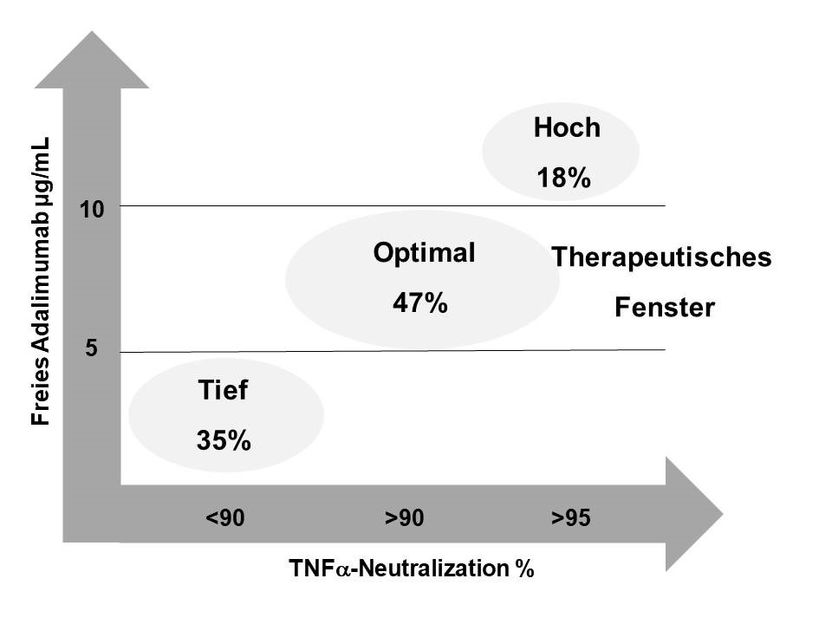
Adjustment of drug dose for rheumatoid arthritis by diagnostic method - Determination of the therapeutic window of adalimumab
Ovarian_cancer
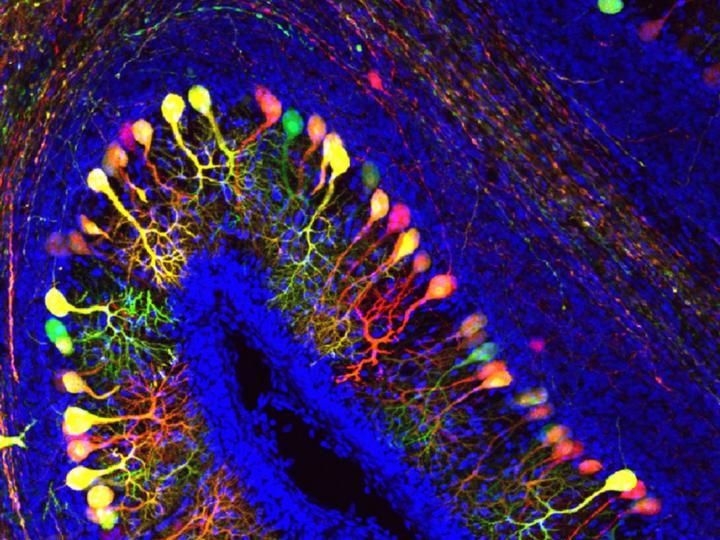
Transporter of thyroid hormones crucial for embryonal brain development
List_of_notifiable_diseases
Scientists find key to regenerating blood vessels
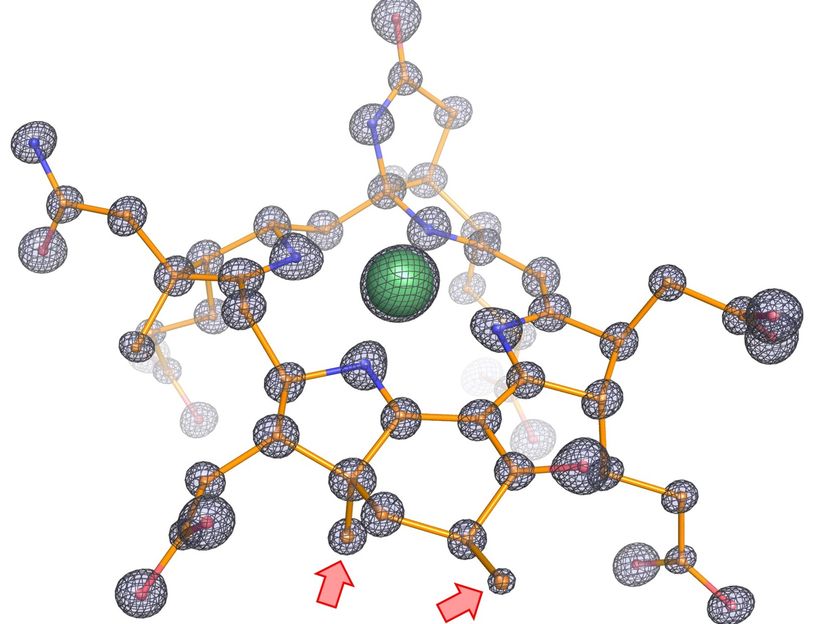
How ethane-consuming archaea pick up their favorite dish - Scientists sucessfully decoded the structure of the enzyme responsible for ethane fixation

Plastic pollution harms bees: risks for global food security - A new review study is the first to systematically show the harmful effects of nano- and microplastics on bees and other beneficial insects
Williams_syndrome
List_of_subjects_in_Gray's_Anatomy:_V._Angiology
QIAGEN and Institute for Animal Health Enter Partnership in Bluetongue Testing























































