Scientists seek to engineer chatter among cells
Communities of cells communicate to coordinate their actions, but how do they coordinate with other communities? Scientists at Rice University and the University of Houston are preparing to find out.
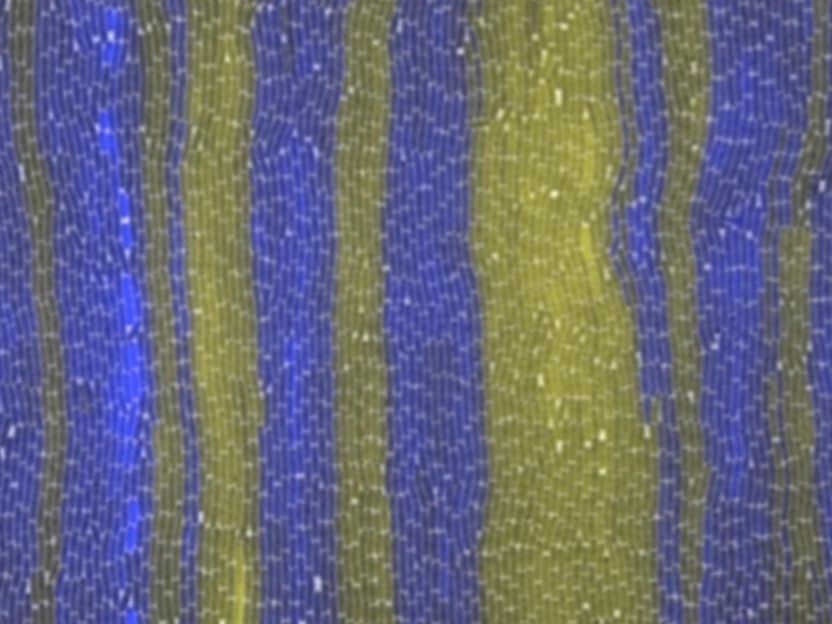
Two strains of bacteria with fluorescent yellow and blue tags organize themselves into an alternating pattern in an experiment at Rice University. Scientists at Rice and the University of Houston have won National Science Foundation grants to study how communities of cells communicate and coordinate. The work could lead to synthetic colonies that can help cure or manage diseases, among other tasks.
The Biodesign Lab/Rice University
Rice synthetic biologist Matthew Bennett and University of Houston mathematician Krešimir Josić are looking into the large-scale dynamics of synthetic microbial consortia to see how they connect. Synthetic microbes are cells in which DNA is modified to carry out a task, like producing a protein on demand or signaling the presence of a target molecule or cell.
They hope to gain insight into how to modify cells -- or build them from the ground up -- to carry out programmed tasks like advancing the production of biofuels or chemicals or adjusting a person's gut microbiome with a synthetic probiotic.
"We want to figure out how to turn simple microbes into multicellular systems that work together, in the same way that cells within animals do," said Bennett, an associate professor of biochemistry and cell biology.
Bennett and Josi? have received combined National Science Foundation grants worth more than $1.5 million to build a framework that describes how cells in large colonies of bacteria communicate with each other and act in concert, even when dispersed among other strains.
The Rice researcher's lab already specializes in building living circuits into bacteria that, for instance, turn protein expression on or off on cue. But these circuits operate at the single- or few-cell level. The next challenge is to design colonies of bacteria that can communicate not only from cell to cell but also across strains to perform computations and make more sophisticated decisions than a single bacterium can.
Their goal is part of a bigger project to model how the elements of life communicate from bottom to top. What the research is leading to, Bennett said, is no less than the ability to program life, either from scratch or in a way that, for instance, allows cooperating bacterial colonies to help cure or manage diseases.
First they have to figure out what happens when many types of cells mix together. The mathematical models, they say, will have to account for signaling among microbes that are moving, growing and dividing as well as co-mingling with other bacterial colonies. "It's going to take a concerted effort to figure out how the emergent properties of multicellular systems come about," Bennett said. "What types of intercellular regulation are required to create nontrivial living structures?
"We're trying to create systems that self-regulate and continue to perform their programmed tasks in the face of challenges," he said. "If a colony is injured or some cells die off, does the remaining colony behave the way it's supposed to? We want to understand how to engineer that kind of robustness."
The researchers also plan to model systems in which cells work best together with their own kind and manage to ignore the signaling cacophony around them. "Our cells are very good at doing what they're supposed to do, even if we get sick or we age," Bennett said. "The pancreas is always going to be the pancreas. It doesn't usually decide to turn spontaneously into a kidney. One goal for larger synthetic multicellular systems is to mimic this ability, called tissue homeostasis."
One challenge to their work is the vast differences in size between the phenomena that generate large-scale multicellular structures and those structures themselves.
"The mechanisms that bring about multicellularity arise from molecules like proteins and DNA that are miniscule compared with the organisms they generate," Bennett said. "Therefore, mathematical models that describe these systems must span length scales that are different by many orders of magnitude, from molecules to large multicellular systems. We have to be able to understand the mathematics behind going from small-scale to large-scale systems.
"There are many ways people use mathematical models to understand cellular life, but they tend to focus on either the very small or the very large," Bennett said. "We want to be able to reconcile those models to generate a better understanding of multicellularity."