RNA Molecules Live Short Lives
Advertisement
A research group at the Biozentrum, University of Basel, has developed a new method to measure the half-life of RNA molecules. The study revealed that commonly used methods provide distorted results and that RNA molecules live an average of only two minutes, ten times shorter than previously assumed.
RNA molecules live an average of two minutes before they are eliminated by an exosome.
University of Basel, Biozentrum
RNA molecules are individual transcripts of the cell’s DNA. They transfer the genetic information of the DNA and provide a template for the production of proteins that regulate all the cell’s processes. The small carriers of information are themselves regulated throughout their lifespan, or rather half-life. After being produced, RNA molecules serve as a template for protein production for a limited time, before they are degraded.
To date, there have been two main scientific methods used to measure the half-life of RNA. As the research team led by Prof. Attila Becskei at the Biozentrum, University of Basel, has now discovered, these conventional methods can be quite imprecise and deliver inconsistent results. Becskei's team has found a new method that demonstrates that RNA molecules do not last for an average of 20 minutes but rather only two minutes. This was a challenging endeavor, because no one knew in advance which method yields the correct results”, says Becskei.
The “gene control method” shows: RNA is short lived
Knowing the half-life of RNA is significant for scientific studies on the cell cycle. The whole process of cell division depends on the right amount of proteins being available at the right time. If RNAs are not available in the right concentrations at a given phase of the cell cycle, errors occur.
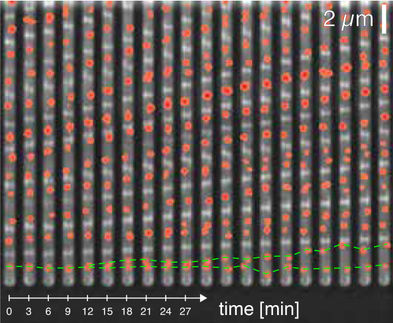
The “gene control method” used by Becskei was already known but has so far not been used to measure the half-life of RNA molecules. This is because this method requires complex genetic engineering and is time-consuming, since only one RNA molecule at a time can be studied. For this a single gene of the DNA is regulated in such a way that the production of RNA can be switched on and off. If the RNA production is stopped, it is possible to measure the survival of the already produced RNA in the cell. From this, the lifetime of this RNA can be determined. “Hence, this method only provides a result for one RNA molecule, but the results are quite accurate”, emphasizes Becskei.
The experiments were repeated for some 50 different genes and showed that 80 percent of all RNAs undergo a rapid turnover, living less than 2 minutes and can be classified as short-lived. Only about 20 percent live longer, for about 5 to 10 minutes. “These results are astounding, if you consider that until now it was assumed that on average RNAs survived 20 minutes in the cell”, says Becskei.
Conventional methods with drawbacks
To date, essentially two main classes of methods have been used by scientists to measure the half-life of RNA molecules. In one method “transcriptional inhibition”, a substance is introduced to the cell, which inhibits RNA production from all genes. “If, however, the production of all RNAs – is inhibited, other processes in the cell are also altered and the cell stops functioning. This distorts the results”, explains Becskei.
The “in vivo labeling” method also has its downside: The RNAs are first labeled and then observed to see how long they can be traced in the cell. However, these labelling with modified molecules can also interfere with cell function and can lead to false results. Thus, all so far used methods have a drawback: The measurement itself influences the processes to be measured. That’s why the results are distorted. “Sometimes it is hard to believe that scientists could have unknowingly worked with methods that produce inconsistent results for almost 30 years”, says Becskei. “It seems that the philosopher of science Paul Feyerabend was right to say that science is often quite anarchistic.”
The research group also compared their method with the existing ones and have found the highest correlation between Becskei's method and a unique variant of the “in-vivo labeling” method. In most cases, both measure classified the same RNAs as stable and unstable even if the mean half-life estimates differ. Now, the team would like to investigate in which areas the latter method provides accurate results and can be used reliably.
Original publication
Antoine Baudrimont, Sylvia Voegeli, Eduardo Calero Viloria,Fabian Stritt, Marine Lenon, Takeo Wada, Vincent Jaquet, Attila Becskei; "Multiplexed gene control reveals rapid mRNA turnover. Advanced Science"; published online July 12, 2017.