Insight into enzyme's 3-D structure could cut biofuel costs
Using neutron crystallography, a Los Alamos research team has mapped the three-dimensional structure of a protein that breaks down polysaccharides, such as the fibrous cellulose of grasses and woody plants, a finding that could help bring down the cost of creating biofuels. The research focused on a class of copper-dependent enzymes called lytic polysaccharide monooxygenases (LPMOs), which bacteria and fungi use to naturally break down cellulose and closely related chitin biopolymers.
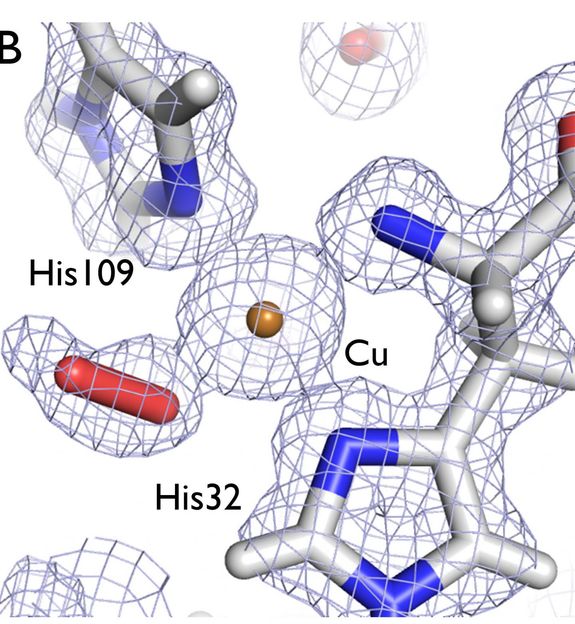
Understanding the structure of an enzyme that helps bacteria break down cellulose and chitin in woody plant fibers can aid in developing better biofuels. In this image, an electron density map (gray) shows the structure of the active site center of the LPMO enzyme under study, depicting a dioxygen molecule (red stick) bound to a catalytic copper ion (bronze).
LANL
"In the long term, understanding the mechanism of this class of proteins can lead to enzymes with improved characteristics that make production of ethanol increasingly economically feasible," said Julian Chen, a Los Alamos National Laboratory scientist who participated in the research.
A multi-institution team used the neutron scattering facility at the Spallation Neutron Source (SNS) at Oak Ridge National Laboratory and the Advanced Light Source (ALS) synchrotron X-ray source at Lawrence Berkeley National Laboratory to study LPMO. Both SNS and ALS are DOE Office of Science User Facilities.
Los Alamos Bioscience Division scientists Chen, Clifford Unkefer, and former postdoctoral fellow John Bacik, working with collaborators at Oak Ridge National Laboratory, Lawrence Berkeley Laboratory, and the Norwegian University of Life Sciences, solved the structure of a chitin-degrading LPMO from the bacterium Jonesia denitrificans (JdLPMO10A).
One of the biggest challenges biofuel scientists face is finding cost-effective ways to break apart polysaccharides such as starches and cellulose, which are widely distributed in plants, into their subcomponent sugars for biofuel production. LPMO enzymes, which are seen as key to this process, use a single copper ion to activate oxygen, a critical step for the enzyme's catalytic degrading action.
While the specific mechanism of LPMO action remains uncertain, it is thought that catalysis involves initial formation of a superoxide by electron transfer from the reduced copper ion. By understanding the location of the copper ion and the constellation of atoms near it, the researchers hope to elucidate more about the enzyme's function. To do this, they rely on first determining the structure of the enzyme.
Although a number of X-ray crystallographic structures are currently available for LPMOs from fungal and bacterial species, this new structure is more complete. The investigators used X-ray crystallography to resolve the three-dimensional structure in clear detail of all the atoms except for hydrogens, the smallest and most abundant atoms in proteins. Hydrogen atom positions are important for elucidating functional characteristics of the target protein and can best be visualized using a neutron crystallography. The investigators used this complementary technique, to determine the three-dimensional structure of the LPMO, but highlighting the hydrogen atoms.
Notably, in this study the crystallized LPMO enzyme has been caught in the act of binding oxygen. Together with the recent structures of LPMOs from a wide variety of fungal and bacterial species, the results of this study indicate a common mechanism of degrading cellulosic biomass despite wide differences in their protein sequences. This study has furthered insight into the mechanism of action of LPMOs, particularly the role of the copper ion and the nature of the involvement of oxygen.
Biofuels research is part of the Los Alamos National Laboratory's mission focus on integrating research and development solutions to achieve the maximum impact on strategic national security priorities such as new energy sources.
Original publication
John-Paul Bacik, Sophanit Mekasha, Zarah Forsberg, Andrey Y. Kovalevsky, Gustav Vaaje-Kolstad, Vincent G. H. Eijsink, Jay C. Nix, Leighton Coates, Matthew J. Cuneo, Clifford J. Unkefer, and Julian C.-H. Chen; "Neutron and Atomic Resolution X-ray Structures of a Lytic Polysaccharide Monooxygenase Reveal Copper-Mediated Dioxygen Binding and Evidence for N-Terminal Deprotonation"; Biochemistry; 2017