Biophysicists say iodine is the solution of biomolecule structures
Advertisement
An international team including researchers from MIPT has shown that iodide phasing--a long-established method in structural biology--is universally applicable to membrane protein structure determination. Knowledge of these structures enables a molecular-level understanding of the workings of eyesight and smell, as well as the nervous and cardiovascular systems.

A slider is shown.
MIPT Press Office
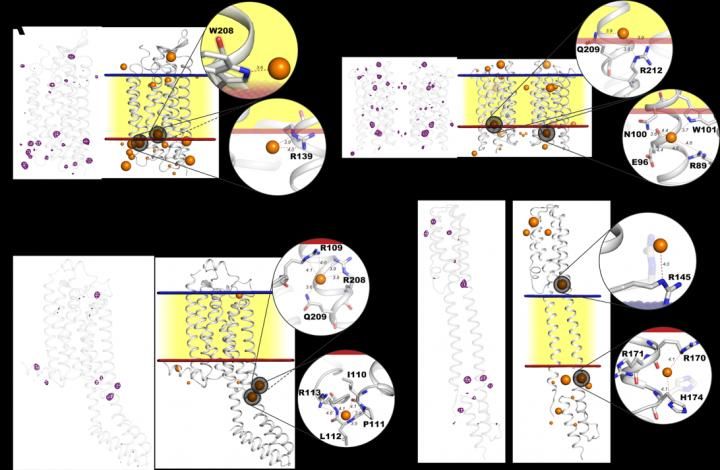
Iodide binding sites in various protein structures. Panels A, B, C, and D show different protein structures with traces of iodide ions depicted in purple (on the left). The same structures are shown embedded in the cell membrane (right). The image illustrates the tendency of iodide ions (orange spheres) to attach to proteins at the outer (blue) or inner (red) surface of the membrane, where positive charge is concentrated.
The study under discussion published in Science Advances

The authors of the study applied the established method of iodide phasing to four membrane proteins representing different classes and discovered that iodide (an ion of iodine) interacts with all four in the same way. This means the method is guaranteed to be successful in uncovering new protein structures vital for pharmaceutics. Because iodide phasing is easy and fast, it can accelerate computer-aided drug development and make it cheaper.
Membrane proteins: the customs service of a cell
All living things are known to be composed of cells. The cells that make up any organism, be it a bacterium like E. coli or a human being, share a common structure. In particular, all cells are surrounded by a protective cell membrane that blocks the passage of the molecules of most substances. Enclosed by a membrane, a cell can maintain internal conditions necessary for complex biochemical processes to run smoothly. However, the survival of a cell also depends on its ability to monitor changes in its external environment and react to them. This is why the genome of every cell encodes several hundreds of proteins of a special kind which are responsible for the cell's interaction with its surroundings. Since these proteins are embedded in the cell membrane, they are referred to as membrane proteins. Another one of their functions is to move into the cell those molecules that are blocked by the membrane of the cell but are nevertheless necessary for its nutrition or biochemical reactions.
Crystallography goes a long way in structural biology, but there is a certain "phase problem"
The biggest breakthrough in structural biology is associated with the Nobel Prize-winning discovery of the double-stranded structure of DNA by Watson and Crick in 1953. The elegant model created by the two scientists was based on prior structural research conducted by their colleague Rosalind Franklin. The double-stranded structure of the DNA molecule (aka the double helix) successfully accounted for the transfer of genetic information between cells and laid the foundation for modern biology.
Crystallography is the chief method employed by structural biology. It enables researchers to elucidate the structure of biomolecules (these are usually proteins) with atomic resolution. Such precision means it is possible not only to look into protein operation at the basic level but also to model their behavior using physical laws.
Crystallography relies heavily on the physical phenomenon called diffraction. To collect diffraction data, a protein crystal is exposed to an X-ray beam. Because molecules in the crystal are in a highly ordered configuration, the X-ray beam diffracts into numerous directions with its intensity amplified manyfold. These diffracted beams can then be picked up and interpreted as a signal, whereby their intensity is measured. However, only averaged signal values for a given direction are available, and part of the information is lost. This is known as the phase problem: The loss phases, i.e., the delay of one signal relative to another, are required for determining the structure of a molecule using diffraction data. Picking up a signal that lacks phases is a little bit like looking at a black-and-white copy of a color image: You can perceive the intensity of every individual point, but the colors are not there and so most of the information is lost.
Phase recovery
Because many crystal structures have already been solved, it is possible to recover the lost phases in studies of new molecules using computer-aided techniques. This involves inferring phases from a known structure and refining them by hand. However, this approach often yields no satisfactory results, particularly in the case of low-resolution data typical for membrane proteins or completely new structures that are not quite like anything that has been solved. Therefore, a different technique is often preferred to deal with these crystals. It is based on the phenomenon of anomalous diffraction, i.e., a certain asymmetry in the diffraction signals produced by heavy elements such as iodine, gadolinium, bromine, and sometimes sulfur. For this method to work, the heavy elements need to strongly bind to the protein molecules in the crystal. This ensures that their atoms are as strictly ordered as the protein molecules themselves to produce a strong diffraction signal. It often takes a long time before the right heavy element is found by trial and error, and many valuable protein crystals are used up in the process.
The researchers showed that anomalous scattering is guaranteed to work if membrane proteins are treated with iodide ions in solution. This is made possible by a common feature of every membrane protein that exists in nature: They are all structured in such a way that there is an extra positive charge on the membrane-solution interface compensating the negative charge on the membrane surface. Iodide interacts strongly with these charges and is selectively attracted to very specific spots on the protein called binding sites, ensuring the success of experimental phase recovery.
"In our study, we demonstrated a successful solution of the structures of four proteins already known from earlier research. The proteins, which come from different organisms, are the following: the light-driven sodium pump from the marine bacterium Krokinobacter eikastus, a fragment of histidine kinase from the Escherichia coli bacterium, a human adenosine receptor, and the proton pump from the marine Actinobacteria clade. With each of the four structures, it can be seen that iodide ions actually bind to the positively charged amino acids at the sites where a protein protrudes from the membrane. Compared to bromide which is also sometimes used to address the phase problem, iodide is more reliable giving more precision in the diffraction measurements," says Igor Melnikov of the European Synchrotron Radiation Facility, the first author of the study, an MIPT graduate.
Original publication
Melnikov, Igor and Polovinkin, Vitaly and Kovalev, Kirill and Gushchin, Ivan and Shevtsov, Mikhail and Shevchenko, Vitaly and Mishin, Alexey and Alekseev, Alexey and Rodriguez-Valera, Francisco and Borshchevskiy, Valentin and Cherezov, Vadim and Leonard, Gordon A. and Gordeliy, Valentin and Popov, Alexander; "Fast iodide-SAD phasing for high-throughput membrane protein structure determination"; Science Advances; 2017