Same but different
Researchers uncover a mechanism of how bacteria with the same genotype can show a different phenotype
Advertisement
Bacterial populations pose an intriguing puzzle: in so-called isogenic populations, all bacteria have the same genes, but they still behave differently, for example grow at different speeds. Researchers at the Institute of Science and Technology Austria (IST Austria) now solved a part of this puzzle by studying how the bacterium Escherichia coli divides up a protein complex that detoxifies cells by pumping multiple drugs such as antibiotics out of the cell. They found that when a bacterium divides into two, the pump proteins are distributed so that one cell inherits more of the pump proteins than the other cell. Bacterial cells that inherit more pump proteins grow more quickly in low concentrations of antibiotics. Partitioning the pump protein is one way to achieve different behaviors in a bacterial population, and could be a stepping-stone to developing antibiotic resistance. The study, published in Science, was carried out by an interdisciplinary duo: an experimentalist, Tobias Bergmiller, and a theorist, Anna Andersson, from the groups of Călin Guet and Gašper Tkačik at IST Austria.
Calin Guet
A bacterial population is made up of thousands of individual bacteria, all with the same genetic make-up. But these individuals can have a range of phenotypes, i.e. look and behave differently, due to random processes in the cells that influence how the genetic instructions are used. For proteins in the cytosol, the liquid inside the cell, these random molecular processes include differences in the break-down of proteins, or random partitioning into the two cells that form during cell division. In the current study, the researchers looked at how a protein complex in the cell envelope, which surrounds the cell, contributes to this heterogeneity or diversity of phenotypes.
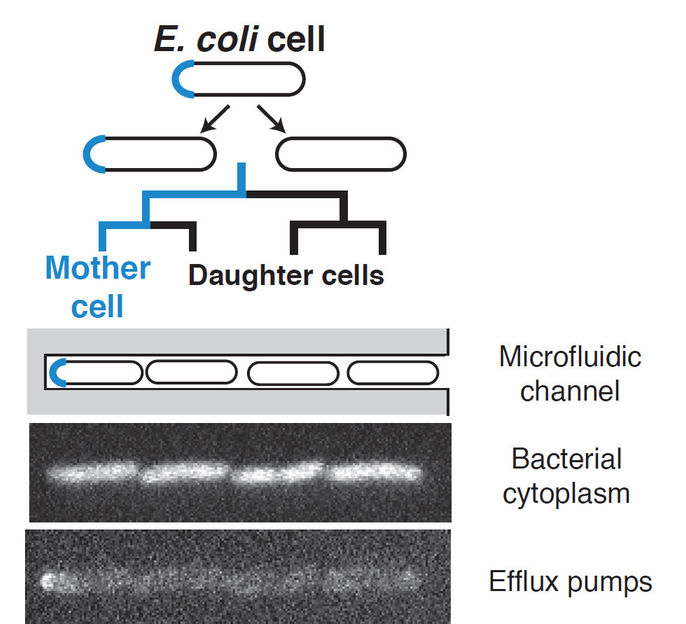
AcrAB-TolC, the protein complex studied, is the main protein complex that pumps drugs out of Escherichia coli cells. It is found in the cell envelope, where proteins cluster together as “islands”. Unlike in the cytosol, where proteins are generally well-mixed by diffusion, proteins in the envelope can segregate asymmetrically. Some of these clusters form at the cell poles, the rounded ends of rod-shaped bacteria like Escherichia coli. In these bacteria, the cells that are “born” at cell division can be distinguished by their poles. One cell, the mother cell, keeps an old cell pole that originated in a past division, where efflux pump proteins have formed stable clusters.
The authors show that AcrAB-TolC accumulates at these old cell poles in mother cells. These mother cells can pump out drugs more efficiently than the daughter cells which did not inherit old cell poles. At low concentrations of antibiotics, the mother cells grow faster than daughter cells. Based on these observations, the researchers developed a mathematical model to identify how this biased partitioning of the drug pump affects the bacterial population. They found that mother cells which can pump out drugs quickly become more common. The population becomes heterogeneous as this extreme phenotype becomes more prevalent as this heterogeneity is long lived.
Călin Guet explains how biased partitioning may be one step on the road to antibiotic resistance: “Recent research by other groups has shown that mutations that cause antibiotic resistance can emerge quickly at low concentrations of antibiotics. Such low concentrations are increasingly found in natural habitats or in patients during drug treatments. Indeed, studies looking at selection due to antibiotics have found that AcrAB, the genes we studied, were amplified. Heterogeneity in a bacterial population that arises through a mechanism of biased partitioning of drug efflux pumps, as we identified in our study, could be a stepping-stone on the path of bacterial populations towards antibiotic resistance.”