The tightest non-aminoglycoside ligand for the bacterial ribosomal RNA A-site
A research group at Tohoku University has made a significant discovery with positive implications for the development of bacteria-fighting drugs. The aminoacyl-tRNA site (A-site) of the 16S RNA decoding region in the bacterial ribosome looks promising for a new era of antibiotic drug development.
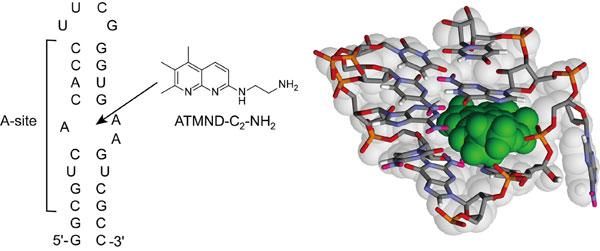
Chemical structure of ATMND-C2-NH2 and sequence of the bacterial (Escherichia coli) A-site-containing RNA model used in this study. It also shows the possible structure of the complex between ATMND-C2-NH2 (green color) and internal loop of A-site.
Seiichi Nishizawa
Traditional aminoglycoside antibiotics are problematic given their high toxicity and the potential for resistance development. The research at Tohoku University focused on bacterial A-site binding small ligands whose structures are distinct from the aminoglycoside family, which offer potential for the development of novel drugs that treat bacterial infections with a reduction in the problems associated with traditional antibiotics.
The research group led by Dr. Seiichi Nishizawa and Dr. Yusuke Sato (Department of Chemistry, Graduate School of Science) has reported a novel small ligand, ATMND-C2-NH2 that has the tightest binding affinity for the bacterial A-site among the non-aminoglycoside ligands.
ATMND-C2-NH2 shows a significant fluorescent quenching response upon selective binding to the internal loop of the bacterial (Escherichia coli) A-site-containing model RNA. ATMND-C2-NH2 has also proven useful as an indicator for assessing ligand/A-site interactions.
The results obtained by the research group offer a rational basis for the generation of novel A-site binding ligands with a view toward novel antibiotics with less toxicity and minimum resistance development.
Original publication
Most read news
Original publication
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.