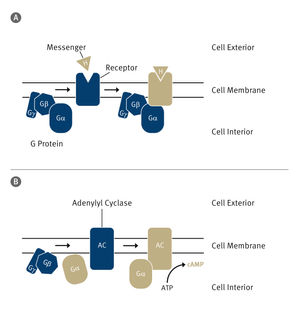
The role of the tunnel
The targeted incorporation of proteins into the membrane is a vital process for cell maintenance; these membrane proteins ensure the proper functioning of the cell’s metabolism, communication with its environment, and energy supply. Protein-sorting mechanisms ensure that membrane proteins are specifically recognized among thousands of different proteins – and are sent to the membrane, where they’re needed. A team headed by Kärt Denks, a doctoral candidate in Professor Hans-Georg Koch’s working group at the Institute of Biochemistry and Molecular Biology at the University of Freiburg, describes this molecular mechanism in detail, using the gut bacterium Escherichia coli. The researchers showed that the signal recognition particle (SRP), present in all living organisms, identifies correct proteins already during their synthesis.
Proteins are synthesized on ribosomes, functional units within the cell, which release proteins via a tunnel to the inner part of the cell. They are then sorted according to a pattern: Proteins to be transported contain an amino acid sequence which serves as a recognition signal for cellular sorting complexes. SRP is one of these complexes. It occurs in bacteria and in organisms with nucleated cells, and is responsible for the recognition of membrane proteins. From earlier investigations, the researchers knew that SRP recognizes membrane proteins even before they are fully synthesized. But there was debate over exactly when. At first it was assumed that the signal sequence had to have emerged completely from the ribosome protein tunnel for the membrane protein to be recognized. But subsequent work indicated that identification took place long before the signal sequence left the ribosome. The new Freiburg research confirms this.
The researchers used a technique which enabled them to examine the contacts between the ribosome and SRP right down to the level of individual amino acids – the very building-blocks of proteins. The team showed that SRP scans the ribosome protein tunnel to find potential substrate proteins. When it recognizes a protein of the right kind, it retracts to the end of the tunnel and positions its binding pocket in order to form a stable complex with the membrane protein. Once it has done that, the SRP begins the process of moving the synthesizing ribosome to its target site at the membrane: where it binds to protein transport channels in order to anchor the protein into the membrane. If this early-recognition fails – if for instance the contact points between the SRP and the ribosomal tunnel have been genetically modified – membrane proteins pile up because they cannot be correctly positioned in the membrane. This leads to cell-division defects.
The research reveals a new complexity in the interaction between ribosomes and protein-sorting complexes: the ribosomal tunnel, long regarded as a passive tube, plays a key role in the coordination of processes which begin during the synthesis of proteins.
The protein-sorting complex SRP scans the ribosome protein tunnel where proteins are being synthesized. When it recognizes a protein of the right kind, SRP positions its binding pocket at the end of the tunnel, where it forms a stable complex with the protein and transports it to the target site in the membrane.
AG Koch
Original publication
Most read news
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.
Most read news
More news from our other portals
Last viewed contents

Unlocking the opium poppy's biggest secret
Category:Industrial_breathing_sets
Margaret_Morse_Nice
Allometric_law

Ingestion of taurine delays aging - Can the aging process be slowed down by raising taurine concentrations to a “young” level?

Excelitas Technologies Acquires Heraeus Noblelight

Ilke Panzer appointed new Greiner Bio-One Division Manager - "We were convinced by her impressive career at major US companies such as General Electric and Johnson & Johnson"
Research combines graphene and painkiller receptor

Molecular machine in nano cage - What a toy: a tiny gyroscope that would fit in a human cell and that can be controlled from the outside

108Labs is building the world’s first cell cultured human milk factory
Was the secret spice in primal gene soup a thickener?