Charting the anatomy of three molecular channels
Using a state-of-the-art imaging technology in which molecules are deep frozen, scientists in Roderick MacKinnon 's lab at Rockefeller University have reconstructed in unprecedented detail the three-dimensional architecture of three channels that provide a path for specific types of ions to travel across a cell's protective membrane. Because such ions are central to biochemical messaging that allows cells to communicate with one another, the findings have implications for understanding how muscles contract, how the heart maintains its rhythm, and many other physiological processes.
This is a switch to calm excited neurons.
Laboratory of Molecular Neurobiology and Biophysics/The Rockefeller University

This image shows how a response is reversed.
Laboratory of Molecular Neurobiology and Biophysics/The Rockefeller University
This image shows a subtle change, another way of working.
Laboratory of Molecular Neurobiology and Biophysics/The Rockefeller University
In all cases, researchers used equipment at the Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center to capture and compile images of the molecules while they were frozen in a thin layer of ice. With this data, they determined the channels' structures in three dimensions, down to the level of individual atoms.
"Structural biology has experienced a true revolution thanks to devices capable of directly detecting the electrons used to visualize a sample," says senior author Roderick MacKinnon, John D. Rockefeller Jr. Professor and head of the Laboratory of Molecular Neurobiology and Biophysics . "With this technology, it has become possible to determine high resolutions structures based on images of individual, randomly oriented protein molecules." Having the structures in hand makes it possible for researchers to investigate long-standing questions about how these molecules function, producing new insights into their biology and potentially aiding the development of treatments for a number of disorders.
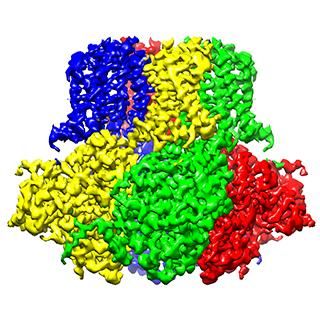
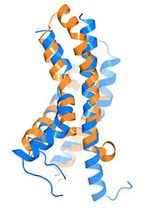
The chloride ion channel: A subtle change, another way of working
The chloride channel, known as CLC, opens to passively allow ions through. However, it has a close relative that moves chloride another way: by exchanging it for protons. In their structural data, Eunyong Park, a postdoc in the MacKinnon laboratory, and Ernest B. Campbell, a research specialist, found a detail that helps to explain how these two similar molecules work so differently: the position of a loop within the pore through which the ions travel. In the exchanger molecule, this loop was already known to partially block the ions' path. In the new CLC structure (above), they saw this loop flipped down, allowing chloride to travel more freely.
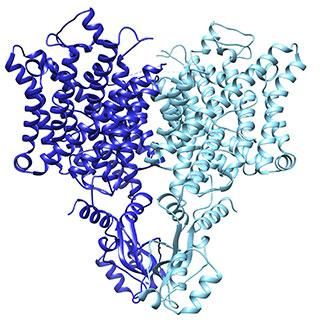
The "pacemaker" channel: How a response is reversed
By allowing potassium and sodium to travel through the cell's membrane, the HCN channel contributes to rhythmic electrical signals, including the pacemaker current within the heart. Among similar channels, HCN's contrarian responses to changes in voltage set it apart. While others open as the cell ramps up for a signal, HCN closes. And unlike the others, it opens as the cell returns to rest. Postdoc Chia-Hsueh Lee found features that contribute to this difference, including an extra-long arm within HCN's voltage detecting sensor (blue) as compared to that of a related potassium channel (orange). The longer arm likely stabilizes the pore in a closed position after the start of an electrical impulse. This and Lee's other findings were detailed January 12 in Cell.
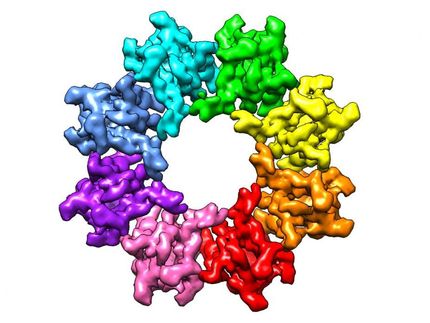
Slo2.2: A switch to calm excited neurons
To prevent high frequency electrical impulses from running out of control, the channel Slo2.2 puts on the breaks by allowing potassium out of the cell. It does so in response to the sodium that rushes in during a signal. Richard Hite, a postdoc, and his colleagues had already determined what Slo2.2 looks like when it is closed, without sodium around. In new research Hite exposed the channel to varying concentrations of its trigger ion, so as to determine the distribution of all the structures that occur simultaneously at a particular sodium concentration--the first such experiment. It turned out that the channel exists only in two conformations, closed and open. As a result, it undergoes a sharp transition when opening, akin to a light switch being turned on.
MacKinnon is a Howard Hughes Medical Institute investigator and a recipient of the 2003 Nobel Prize in Chemistry.