Manipulating signals in bacteria could reduce illnesses
Advertisement
The University of Illinois at Chicago College of Pharmacy has received a five-year, $1.25 million federal grant to continue its research into how bacteria that cause streptococcal infections can be manipulated.
By studying the chemical signals of bacteria, Michael Federle, associate professor of medicinal chemistry and pharmacognosy, is learning how bacteria cells communicate with one another. The research goal is to convince sickness-causing microorganisms to remain in a nonhostile state.
Every antibiotic to fight bacteria has in some instance seen a method of resistance develop against it, he said.
"In recent years, the urgent need to identify new alternatives to antibiotics that fight bacterial infections has inspired the concept of inhibiting fundamental mechanisms of pathogenesis, rather than inhibiting processes essential to bacterial growth," Federle said.
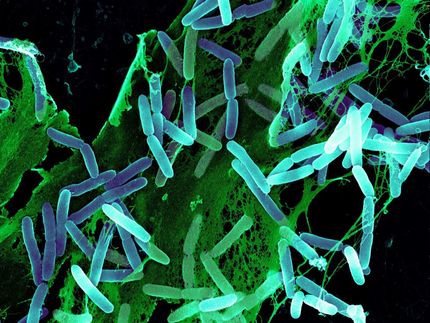
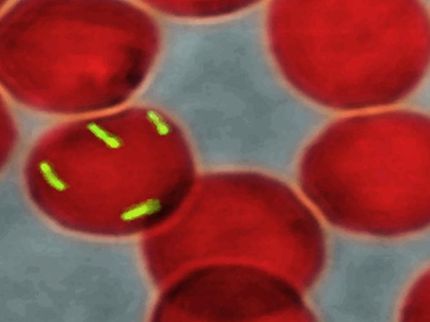
Federle's research centers on streptococci, a group of bacteria responsible for infections such as strep throat, meningitis, bacterial pneumonia, endocarditis, erysipelas, and the "flesh eating" disease, necrotizing fasciitis. He is exploring ways to disrupt biofilms -- communities of bacteria that live on a surface that are extremely resistant to antibiotics.
Federle and his colleagues have studied the molecular mechanisms of the signaling pathways in streptococci. The bacteria use small peptides -- a chain of amino acids that are the building blocks of protein -- that are secreted into the bloodstream and can turn on different genes. Such peptides function as pheromones and can turn on the genes that may contribute to illnesses.
"We found several examples of these peptides, which are detected by receptors of a family of proteins called Rgg," Federle said. "We know they turn genes off and on, but we're expanding our research to discover what exactly their functions are.
"We're trying to find out if the signals -- the small pheromones -- of all the bacteria can be manipulated. If the bacteria are using these signaling pathways to colonize, that contributes to making a person sick. We want to see if they can be manipulated so they won't do that."
Federle's latest research will focus on understanding how pheromones interact with the Rgg proteins and how those proteins interact with DNA. He is also working to identify small molecules and peptides that specifically inhibit Rgg function. Determining the mechanistic basis of inhibitor function and identifying the structural features of those compounds will guide anti-infective drug development in the future, Federle said.
"If we can manipulate bacteria by understanding the chemical signals they use, then we can interfere with the bacteria's ability to make people sick," he said. "We'll try to fool the bacteria by artificially stimulating them."