TET proteins drive early neurogenesis
The fate of stem cells is determined by series of choices that sequentially narrow their available options until stem cells' offspring have found their station and purpose in the body. Their decisions are guided in part by TET proteins rewriting the epigenome, the regulatory layer of chemical flags that adorn the genome and influence gene activity, report researchers at La Jolla Institute for Allergy and Immunology and UC San Diego.
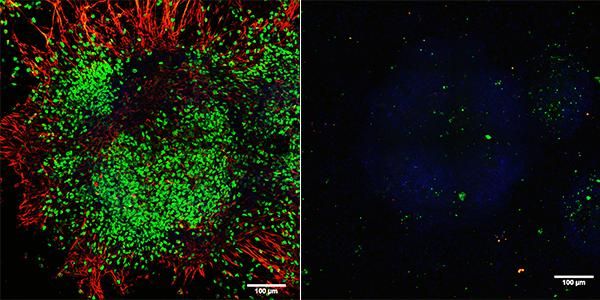
TET proteins helps steer stem cells toward the neuronal lineage during early embryogenesis. Left: Control cells express neuronal markers such as Sox1 (shown in green) and Tuj1 (shown in red). Right: In the absence of TET proteins, stem cells fail to express any neuronal markers. Both: Cell nuclei are shown in blue.
Courtesy of Dr. Xiang Li, La Jolla Institute for Allergy and Immunology.
A study led by LJI professor Anjana Rao, Ph.D., details how TET proteins help stem cells strike the proper balance between developing into neural tissue or taking the heart and muscle route at an early and crucial intersection.
"In particular, TET proteins are important to steer stem cells toward the neural lineage during early embryogenesis," says postdoctoral researcher and first author Xiang Li. "It confirmed what we had suspected--that TET proteins help drive early neurogenesis--but until now the formal proof had been missing." As a clearer picture of the molecular signals triggering certain cell fate decisions emerges, Li hopes that "it brings us closer to using stem cells to treat neuronal disease."
TET proteins influence gene expression by specifically modifying methyl groups attached to cytosine, one of the four letters in DNA. Although these methyl group alterations are important in their own right, they also facilitate the complete removal of methyl groups from cytosine. While methylation and the enzymes required to add methyl groups have been extensively studied as a fundamental force that that helps guide stem cells toward their future lineage during embryonic development, much less is known about the reverse process.
"During early embryogenesis waves of demethylation sweep the genome but we haven't really understood what the relevance is," senior author Sylvia Evans, Ph.D., a professor in the Skaggs School of Pharmacy and Pharmaceutical Sciences at UC San Diego, who collaborated with Rao's team. "By creating triple knockouts that miss all three genes coding for TET proteins and studying them, in vivo, we have learned that yes, the dynamic modulation of the methylation status is critically important for driving developmental gene expression programs in the early embryo."
Li started his investigations with mouse embryonic stem cells (mES) deficient for Tet3 only, or deficient for all three members (Tet1/2/3) of the Tet protein family. During differentiation, cell types from both mouse strains steered clear from committing to the neuroectodermal lineage, which goes on to form the brain and spinal cord and is considered the "default" pathway for mECs cultured under certain culture conditions.
A genome-wide analysis revealed that Tet3 exerts its effect through the inhibition of Wnt-signaling. Wnt signalling is necessary for the proper formation of heart and muscle. "Without Tet3, Wnt signaling is no longer suppressed and can push differentiating stem cells toward a mesodermal cell fate," explains Li. The mesoderm develops into connective tissue, muscle, bone, as well as the urogenital and circulatory system including the heart.
The results from lab-grown mECs prompted the researchers to look for developmental abnormalities in Tet3 and Tet1/2/3-deficient mice. The Tet3 knock-out showed no overt cardiac or neuronal abnormalities, which pointed to functional redundancies between different Tet family members. Tet1/2/3-deficient mice, however, had major defects in neural development, confirming a key role for TET proteins in guiding stem cells down the right path.