Multi-institutional collaboration uncovers how molecular machines assemble
Ribosomes--macromolecular machines consisting of RNA and proteins that twist, fold and turn--are responsible for making all of the protein within a cell and could hold the key to deciphering a range of diseases. Despite the intricacies of ribosomes, cells are able to churn out 100,000 of them every hour. But because they assemble so speedily, researchers haven't been able to figure out how they come together.
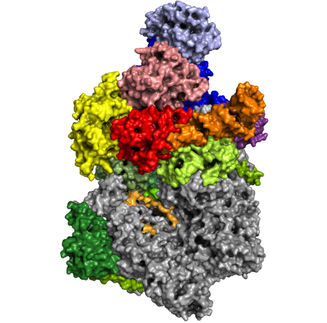
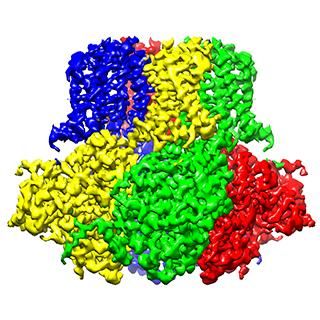
Ribosomes churn out new proteins within a cell and are, themselves, intricate blends of RNA and protein. A multi-institutional collaboration has now solved the structure of a crucial piece of the ribosome as it assembles. This new study shows that it will be possible to examine other large, dynamic molecules with new high-resolution imaging methods.
Salk Institute
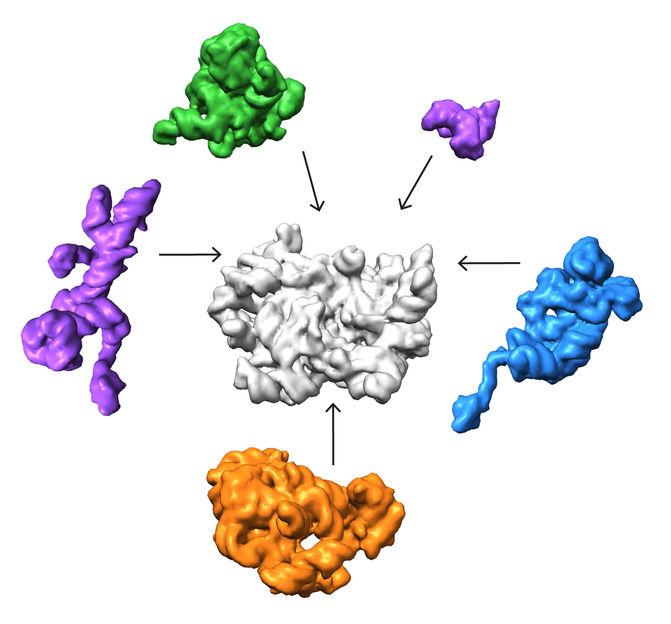
A collaboration led by the Salk Institute and The Scripps Research Institute in La Jolla, California, deployed a cutting-edge imaging method called single-particle cryo-electron microscopy (cryo-EM) and accompanying analysis tools to decipher some of the key steps for how ribosomes are assembled, a first step in understanding their roles in health and disease.
Salk Institute
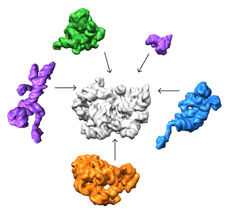
A collaboration led by Salk Institute for Biological Studies and The Scripps Research Institute in La Jolla, California, deployed a cutting-edge imaging method called single-particle cryo-electron microscopy (cryo-EM) and accompanying analysis tools to decipher some of the key steps for how ribosomes are assembled, a first step in understanding their roles in health and disease.
"These new structures we captured with cryo-EM show that it is possible to image and interpret diverse molecular machines in action," says the study's co-senior author Dmitry Lyumkis, a Helmsley-Salk Fellow at Salk. "This is a completely different way of seeing and doing structural biology. This paper is a prime example of the fact that we can do far more intricate analyses than have ever been expected."
Understanding molecular structures is important not only for basic research into biology but also for the drug development process to better understand how to make safer and more effective medicines. Researchers traditionally turn to X-ray crystallography, a method that requires its users to extensively purify a molecule and then re-make it in crystal form, but this method has limitations.
In the past few years, advances in cryo-EM have allowed scientists to image single particles with resolution comparable to that of traditional X-ray methods. But in single-particle cryo-EM, proteins (the "particles") are flash-frozen and imaged using streams of electrons, meaning the molecules don't need to be crystallized and can retain much of their native structure. Although cryo-EM has been around for a while, new cameras are making it easier to capture proteins at high resolution before the electron spray zaps them.
Importantly, computational tools for analyzing cryo-EM data have matured such that researchers can now purify molecules in silico by a computer rather than through traditional biochemical approaches. This becomes a much more powerful approach for separating mixtures of species, allowing the researchers to identify and distinguish structurally distinct populations of particles in greater detail than before.
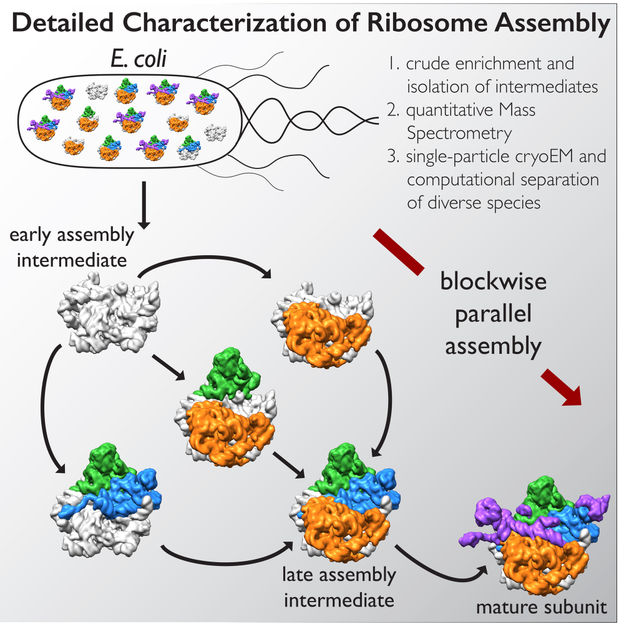
In the new study, co-senior author James Williamson, professor of molecular biology and chemistry at The Scripps Research Institute, and his team developed a method to stall one major component of ribosomes, the 50s subunit, from coming together so quickly. The scientists were able to chemically pause a mixture of different molecules in various stages of assembly.
Lyumkis's group then used high-end cryo-EM to image and analyze these stalled structures, which had not been attempted for such a mix of diverse forms of a particular molecule.
"Others have shown that you can capture a couple of different structural states of a molecule," Lyumkis says. "But, as far as I know, no one has tried to take this crude mixture of stuff, put it onto a cryo-EM grid, and ask what was in there."
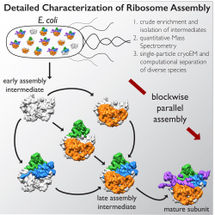
The team found that there are at least 15 types of complexes in the mixture, 13 of which are actively assembling 50s subunits. They imaged each of these structures at a resolution high enough to decipher the protein and RNA constituents. They were then able to use computer algorithms to order the complexes according to their assembly pathway.
The team's analysis suggests that ribosomes can take several different routes for assembly, which is important to ensure that the process is efficient and can withstand a variety of cell stresses, according to Williamson. "If you imagine an assembly line where every step has to happen in sequential order, and there is a problem at one of those steps, everything grinds to a halt," he says. "If there are parallel pathways, then assembly can proceed through other channels until the problem is resolved.
It took the scientists over a year to make sense of the structures, employing relatively new image analysis tools. But they have laid the groundwork to study other large, dynamic, and structurally heterogeneous molecular machines, which, Lyumkis says, will lead to new basic science and translational discoveries.