Personalized antibiotic treatment
Researchers from Freiburg have developed a sensor platform that quantifies antibiotics in human blood within minutes
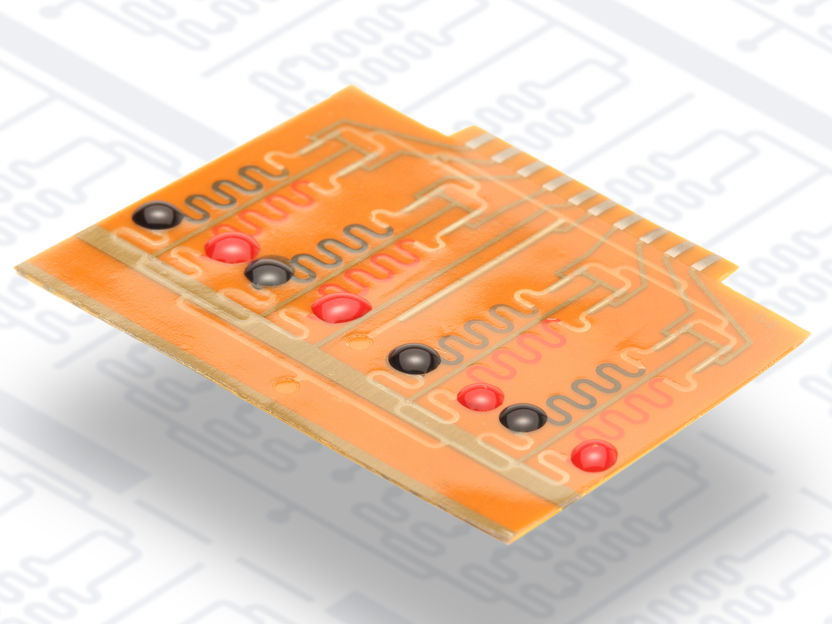
The electrochemical biosensor system for point-of-care testing.
Andreas Weltin
A team of researchers from the University of Freiburg has developed a system inspired by biology that can detect several different antibiotics in human blood or other fluids at the same time. This biosensor system could be used for medical diagnostics in the future, especially for point-of-care testing in doctors’ practices, on house calls and in pharmacies, as well as in environmental and food safety testing. The researchers focused their study on the antibiotics tetracycline and streptogramin in human blood. “The analysis takes only 10 minutes, from sample to result,” said the microsystems engineer Dr. Can Dincer, who is the head of the research team: “Our study was about demonstrating the applicability of the platform.” Based on these findings, the group is currently working on developing a method to determine how quickly the human body breaks down antibiotics, thus enabling the dosage of medications to be adjusted to each patient. “This technology could pave the way for personalized antibiotic treatments in the future,” Dincer said.
The all-too-frequent use of antibiotics in human and veterinary medicine causes pathogens to develop resistance. Multidrug resistant bacteria are the reason for an increasing number of life-threatening infections that are difficult to treat with medications available today. In this context, biosensors have so much potential in research, since they are inexpensive and easy to work with. It is expected that biosensors can be employed to customize antibiotic treatments to fit each patient`s requirements, thereby decreasing the development of resistant bacteria in the future.
The electrochemical biosensor platform was developed by Prof. Dr. Gerald Urban’s research group. It works with extremely small amounts of liquid. “The major advantage of this system is that we can measure up to eight different substances at the same time, quickly and simply,” Dincer said. The researchers combined their chip technology with a method developed earlier by the bioengineering expert Prof. Dr. Wilfried Weber, also from the University of Freiburg. The method is based on a naturally occurring sensor protein in resistant bacteria to recognize antibiotics and activate their defence mechanisms. These bacterial sensors react quickly, sensitively and specifically to antibiotics, which makes them ideal for analytical testing. Essentially, the bacteria are providing the researchers with a tool that can be applied to fight them back in the long-run.
The eight researchers from the University of Freiburg involved in the interdisciplinary study include Lucas Armbrecht, Dr. Can Dincer, Dr. Jochen Kieninger, André Kling, Edvina Qelibari and Prof. Dr. Gerald Urban – all from the Sensors Lab of the Department of Microsystems Engineering (IMTEK) – as well as Claire Chatelle and Prof. Dr. Wilfried Weber from the Synthetic Biology Department of the BIOSS Centre for Biological Signalling Studies cluster of excellence and the Faculty of Biology.
Original publication
Other news from the department science
These products might interest you
Most read news
More news from our other portals
See the theme worlds for related content
Topic world Diagnostics
Diagnostics is at the heart of modern medicine and forms a crucial interface between research and patient care in the biotech and pharmaceutical industries. It not only enables early detection and monitoring of disease, but also plays a central role in individualized medicine by enabling targeted therapies based on an individual's genetic and molecular signature.

Topic world Diagnostics
Diagnostics is at the heart of modern medicine and forms a crucial interface between research and patient care in the biotech and pharmaceutical industries. It not only enables early detection and monitoring of disease, but also plays a central role in individualized medicine by enabling targeted therapies based on an individual's genetic and molecular signature.