A cooperative way to make ammonia
nitrogen is essential for life, but nature's main source of nitrogen is the gas found in our atmosphere -- a gas that does not react easily with other elements. Some specialized bacteria turn the air's nitrogen molecules into ammonia so that it can be used to make proteins -- the building blocks, machines and power plants of cells. A little over a century ago, chemists developed a way of making ammonia industrially out of atmospheric nitrogen, but the man-made process uses a lot of energy.
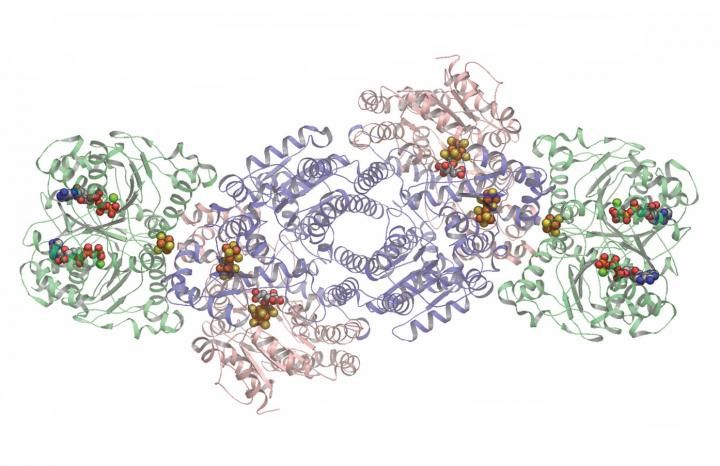
Two copies each of three proteins (in different colors) make up the nitrogenase complex. In this image, the spots where chemistry happens are highlighted as stick-and-ball molecules.
Simone Raugei/PNNL
A better understanding of how bacteria fix nitrogen molecules into ammonia could lead to energy savings in industrial processes such as those that produce fertilizer. Researchers are studying the bacterial enzyme that does this, a complicated enzyme called nitrogenase. In new work, researchers discovered essential information about the manner in which nitrogenase produces ammonia.
"We propose that the enzyme works as a two-stage engine where the two parts work nearly synchronously," said computational chemist Simone Raugei, a coauthor at the Department of Energy's Pacific Northwest National Laboratory. "All this is achieved by complex communication between the two halves."
How nitrogenase's two identical halves worked together had long stumped chemists, but these results reveal that they cooperate in an efficient way. While one half is working to break down a nitrogen molecule, moving some of the pieces in place to make ammonia, the other half is waiting for the first to complete part of it task. Then, the authors hypothesize, the second half starts on a new nitrogen molecule while the first half finishes the later steps, the two ratcheting back and forth in sync until a whole nitrogen molecule is dismantled and rebuilt into ammonia.
Ammonia machine
The working nitrogenase is actually a complex of several proteins and metals, along with energy-storage molecules called ATP to power the whole thing. The complex has two symmetrical halves. Either of these can convert molecular nitrogen to ammonia, molecules that contain one nitrogen atom bonded to three hydrogen atoms. Although researchers know what the complex looks like, many of the details of how it actually works have not been resolved.
Overall, the nitrogenase, burning through ATP, combines atmospheric nitrogen with protons and electrons to build ammonia molecules. The components on either side of nitrogenase do the same thing: some figuratively unbolt the electrons holding the atoms together; others move things around and into place, and others fasten the pieces into ammonia. The halves are identical but symmetrical, positioned relative to each other like yin and yang symbols.
The enzyme produces two molecules of ammonia and one molecule of hydrogen as a byproduct for each molecule of nitrogen dismantled. This process requires eight protons and eight electrons, donated by other components in the bacterium. Each half moves only one electron at a time, so it must repeat the action eight times to make two ammonia molecules.
For a long time, many researchers thought the two symmetrical halves took up electrons independently. But lab experiments suggested that might not be the whole story.
"A kinetic model where the two halves work independently didn't match new data collected by our laboratories," said PNNL's Lance Seefeldt.
If the two halves weren't independent, what exactly was going on? The team, including Seefeldt at PNNL and Utah State University, Raugei, Brian Hoffman at Northwestern University, Edwin Antony at Marquette University and eight other researchers, decided to find out.
To figure it out, the team mapped out three possible ways the enzyme could work, with the plan to see which one fit experimental data best. One way was the commonly accepted version -- both sides worked completely independently, never interfering with each other. In another, only one side worked at a time, the other side being completely shut down until the first side was finished. The third was somewhere in between.
Each method made certain predictions about the various steps in the chemical reactions. Then the team performed the reactions in the lab and crunched the numbers.
This one's just right
They found that the amount of byproducts produced in the experiments was about half expected if the two sides worked completely independently, so they ruled the first model out. And the nitrogenase used up too much ATP for only one side to be working at a time, so they ruled the second model out.
In true Goldilocks fashion, the data worked best with the third model. In this, one side performed a chemical reaction and partially suppressed the activity on the other side.
To visualize what was going on, the team investigated the molecular details of the nitrogenase complex computationally. They saw something rather interesting in their computer simulation.
When one side moved an electron through the nitrogenase machine, the complex shifted in shape. Because the sides are symmetrical, moving one side up moves the other side down. Doing this positions different parts of the machine for different chemical reactions.
Large-scale (for a protein) shape changes like this are a common way for proteins to be turned off and on, but it hasn't often been seen in nitrogenase catalysis, said Raugei.
"Long-distance communication as a way to regulate electron movement is unusual in catalysis," he added.
In future work, the researchers hope to produce a more precise computational simulation to better explore how nitrogenase makes ammonia.
























































