Deep learning applied to drug discovery and repurposing
In a recently accepted manuscript scientists from insilico medicine, Inc located at the Emerging Technology Centers at Johns Hopkins University in collaboration with Datalytic Solutions and Mind Research Network presented a novel approach applying deep neural networks (DNNs) to predict pharmacologic properties of many drugs. In this study, scientists trained deep neural networks to predict the therapeutic use of a large number of drugs using gene expression data obtained from high-throughput experiments on human cell lines. Authors used a sophisticated approach of measuring the differential signaling pathway activation score for a large number of pathways to reduce the dimensionality of the data while retaining biological relevance and used these scores to train the deep neural networks.
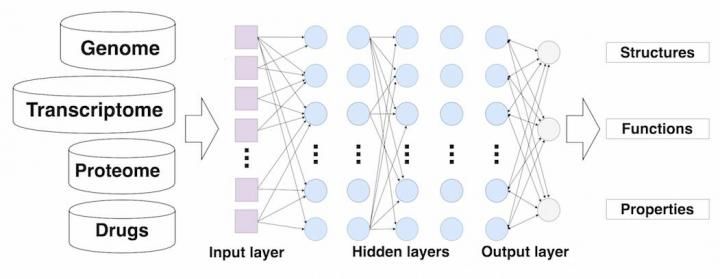
This image shows deep neural networks for drug discovery
Insilico Medicine, Inc.
"The world of artificial intelligence is rapidly evolving and affecting every aspect of our daily life. And soon this progress will be felt in the pharmaceutical industry. We set up the Pharma.AI division to help pharmaceutical companies significantly accelerate their R&D and increase the number of approved drugs, but in the process we came up with over 800 strong hypotheses in oncology, cardiovascular, metabolic and CNS space and started basic validation. We are cautious about making strong statements, but if this approach works, it will uberize the pharmaceutical industry and generate unprecedented number of QALY", said Alex Zhavoronkov, PhD, CEO of Insilico Medicine, Inc.
Despite the commercial orientation of the companies, the authors agreed not to file for intellectual property on these methods and to publish the proof of concept. Insilico Medicine is currently developing multimodal deep neural networks to predict a broad range of properties of drugs, small molecules and natural compounds for a range of applications including treating common and rare diseases, aging, regenerative medicine and increasing response rates in cancer immunotherapy.
"The field of machine learning have recently witnessed an impressive breakthrough in the area of pattern recognition and computer vision. Deep learning, technology to thank for this, continues to disrupt traditional approaches in many other subfields of machine learning. Originally in the 60s, inspired by how the brain works (at least how we understood it back then) deep learning has now developed into a mature engineering concept. The brain however, does not cease to puzzle researchers and, I am sure, contains more sources of inspiration for the future powerful methodologies.", said Sergey Plis, PhD, Director of Machine Learning at the Mind Research Network and CEO of Datalytic Solutions.
In this study scientists used the perturbation samples of 678 drugs across A549, MCF-7 and PC-3 cell lines from the Library of Integrated Network-Based Cellular Signatures (LINCS) project developed by the National Institutes of Health (NIH) and linked those to 12 therapeutic use categories derived from MeSH (Medical Subject Headings) developed and maintained by the National Library of Medicine (NLM) of the NIH. To train the DNN, scientists utilized both gene level transcriptomic data and transcriptomic data processed using a pathway activation scoring algorithm, for a pooled dataset of samples perturbed with different concentrations of the drug for 6 and 24 hours. Cross-validation experiments showed that DNNs achieve 54.6% accuracy in correctly predicting one out of 12 therapeutic classes for each drug. One peculiar finding of this experiment was that a large number of drugs misclassified by the DNNs had dual use, suggesting possible application of DNN confusion matrixes in drug repurposing.
"This study is a proof of concept that DNNs can be used to annotate drugs using transcriptional response signatures, but we took this concept to the next level. We developed a pipeline for in silico drug discovery, which has the potential to substantially accelerate preclinical stage for almost any therapeutic and came up with a broad list of predictions with multiple in silico validation steps that, if validated in vitro and in vivo, can almost double the number of drugs in clinical practice", said Alex Aliper, president of research, Insilico Medicine, Inc and the lead author of the study.