Theory unlocks the secret behind protein-membrane interactions
Understanding cell behavior better by elucidating how proteins work with cell membranes
Trillions of cells -- all different shapes and sizes -- form a human body's structure. Surrounding each cell is a membrane, jointly acting as hostess and security -- welcoming certain information into the cell while making sure its components don't spill out into the body's void. Much is known about how the individual pieces of a cell work, but a significant understanding of how proteins interact with the cell membrane has remained a mystery until now, following a recent study at the University of Missouri.
"When you think about the fundamental components of living systems, proteins are among the most important, right up there with nucleic acids," said Gavin King, an associate professor of physics in the College of Arts and Science at MU, and joint associate professor of biochemistry. "Proteins execute more activity in the cell compared to DNA."
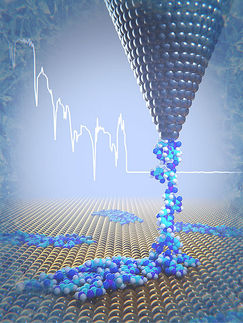
Proteins are the workhorses of a cell. About 30 percent of proteins in any given cell frequently interact with membranes or reside within membranes in order to facilitate and regulate the flow of information and materials in and out of cells. Using high precision atomic force microscopy experiments, King's team measured the force required for proteins to break free from the membrane.
"Imagine you are going fishing, and your fishing rod is a force microscope," King said. "At the end of our fishing rod we attached a lure, or in this case a really short protein. In a very careful and controlled manner, we lower the fishing rod to the vicinity of a membrane. In a way we can't control or directly observe, the lure is frequently bitten by the fish, which in this case is the membrane. When the fish bites, we can pull the lure back and we can ask how much force it takes to pop the lure out of the fish's mouth. What surprised us is that if you do that same experiment repeatedly, you get different results. We were struggling to find a model that could fit this complexity."
To answer this question, Ioan Kosztin, a professor of physics in the College of Arts and Science at MU, partnered with King and developed a theoretical model that shows there is more than one way a protein can break free of the membrane involving several different pathways. They discovered that the protein-membrane interaction can exhibit a "catch-bond" behavior.
"Catch-bond behavior is similar to a Chinese finger trap, where counterintuitively, the harder one pulls on the trap, the stronger the trap pulls back," Kosztin said. "Though similar behavior has been previously described on a cellular level, to our knowledge, this is the first report for protein-membrane interactions."
The researchers hope this discovery will provide a foundation for future studies on signaling pathways in cells and how drugs vary cellular functions.
Original publication
Milica Utjesanovic, Tina R. Matin, Krishna P. Sigdel, Gavin M. King & Ioan Kosztin; "Multiple stochastic pathways in forced peptide-lipid membrane detachment"; Scientific Reports; 2019
Most read news
Original publication
Milica Utjesanovic, Tina R. Matin, Krishna P. Sigdel, Gavin M. King & Ioan Kosztin; "Multiple stochastic pathways in forced peptide-lipid membrane detachment"; Scientific Reports; 2019
Topics
Organizations
Other news from the department science
These products might interest you

Hahnemühle LifeScience Catalogue Industry & Laboratory by Hahnemühle
Wide variety of Filter Papers for all Laboratory and Industrial Applications
Filtration Solutions in the Life Sciences, Chemical and Pharmaceutical Sectors

Hydrosart® Ultrafilter by Sartorius
Efficient ultrafiltration for biotech and pharma
Maximum flow rates and minimum protein loss with Hydrosart® membranes

Hydrosart® Microfilter by Sartorius
Hydrophilic microfilters for bioprocesses
Minimal protein adsorption and high flow rates

Sartopore® Platinum by Sartorius
Efficient filtration with minimal protein adsorption
Reduces rinsing volume by 95 % and offers 1 m² filtration area per 10"

Polyethersulfone Ultrafilter by Sartorius
Reliable filtration with PESU membranes
Perfect for biotechnology and pharmaceuticals, withstands sterilisation and high temperatures

Polyethersulfone Microfilter by Sartorius
Biotechnological filtration made easy
Highly stable 0.1 µm PESU membranes for maximum efficiency

Sartobind® Rapid A by Sartorius
Efficient chromatography with disposable membranes
Increase productivity and reduce costs with fast cycle times

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.
Most read news
More news from our other portals
Last viewed contents
Clitocybe_dealbata
(Bis(trifluoroacetoxy)iodo)benzene
William_Thompson_Sedgwick
Category:Glycolysis
Magnesium_sulfate
Methylprednisolone
George_Heist
Scripps Research scientists crack mystery of protein's dual function - The work provides insight into potential therapeutic for cancer and eye diseases
C3_carbon_fixation
Asperger_syndrome
Veterans_Health_Information_Systems_and_Technology_Architecture