Scientists X-ray protein crystals inside cells
Novel approach to unveil the structure of challenging biomolecules
Scientists from the European Molecular Biology Laboratory EMBL have teamed up with experts from DESY and the U.S. SLAC National Accelerator Laboratory to X-ray protein crystals naturally produced within biological cells. The study at the world's currently most powerful X-ray laser LCLS at SLAC shows that these natural Crystals can be used to determine the spatial structure of the proteins.
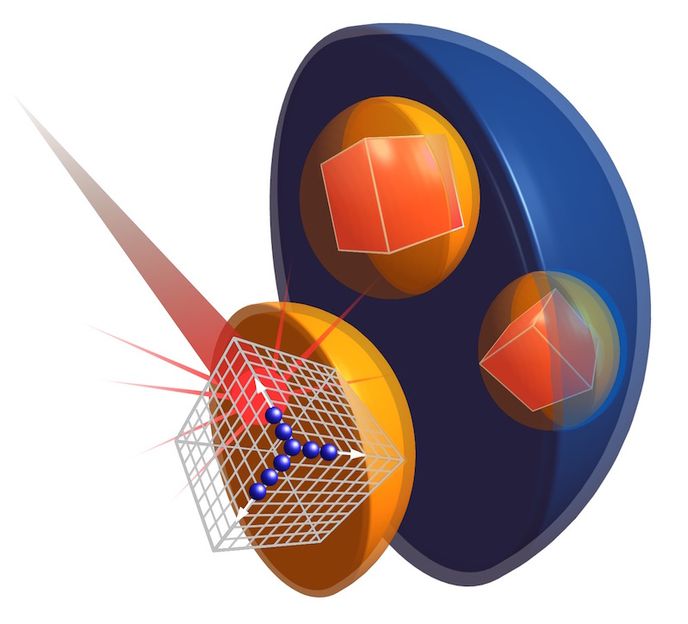
Free-electron laser beam hits a peroxisome containing protein crystals.
Thomas Seine, EMBL/CFEL
Structural biologists strive to unveil the structure of biomolecules, as the form of a protein can say much about its function and may ultimately be used as a basis for the development of new, tailor-made drugs. Crystallography uses X-rays to probe the 3D atomic structure of proteins that have been captured in crystalline form, but the technique has its limitations. In their study published in the International Union of Crystallography Journal (IUCrJ), the team instead used crystals grown inside yeast cells for crystallography experiments at the Linac Coherent Light Source (LCLS).
While structural biologists are familiar with the concept of growing protein crystals in the lab for X-ray crystallography experiments, many may not know that some organisms produce crystals naturally within their cells. “When we heard about these naturally forming crystals, we wondered whether we could use them for crystallography experiments,” says Daniel Passon, a postdoc in the group of Matthias Wilmanns at EMBL's Hamburg branch and one of the lead authors of the study. “Producing protein crystals in the lab for crystallography experiments is not always easy – imagine if we could get cells to do this for us: a tiny crystal factory in a cell!”
The group studied crystals that occur naturally in parts of the cell called peroxisomes. These organelles break down large molecules such as fatty acids, keeping toxic processes safely within their bounds and away from the rest of the cell. In Hansenula polymorpha yeast cells, a protein called alcohol oxidase breaks down methanol molecules into useful byproducts.
To make effective use of the restricted space within the peroxisome, alcohol oxidase molecules are packed tightly into crystals; despite being so densely packed, the enzyme molecules inside this crystal are still active. “We think of crystals as rigid entities, but in fact they are not entirely solid,” explains Arjen Jakobi from EMBL, the other lead author. “The methanol molecules pass through the crystals, reacting with the oxidase to become detoxified.”
While all of these yeast cells have peroxisomes, some cells have more peroxisomes than others, and peroxisome size varies from cell to cell, too. This natural variation posed a problem, as the researchers needed the crystals to be as large as possible, and as similar to each other as possible.
The researchers worked closely with colleagues at the University of Groningen who identified a mutant strain of yeast that only produces one large peroxisome per cell, each containing one large crystal. “Large is of course relative,” says Passon. At 0.001mm, analysis of the crystals required the enormous power of an X-ray Free-Electron Laser (XFEL) like LCLS. “Free-Electron Lasers produce a large amount of photons in small bursts and have a very small parallel beam,” explains Wilmanns, Head of EMBL Hamburg, who oversaw the research. “This makes them ideal for looking at such small crystals.”
The experiment was novel not only for EMBL’s scientists. “This field is its infancy and there are few leading experts worldwide,” says Wilmanns. “We teamed up with the Coherent Imaging Division at the neighbouring Center for Free Electron Laser Science (CFEL) at DESY and University of Hamburg, and benefited from the considerable experience and expertise of division Director, Henry Chapman and his group” he adds.
Having done some initial validation experiments on the beamlines at DESY in Hamburg, Wilmanns, Chapman and their teams set off to SLAC with their precious peroxisomes. The group prepared two types of samples for the experiment: one with the peroxisomes inside their cells and the other with just the peroxisomes, removed from cells. “Surprisingly, we got better data when we measured the peroxisome inside the cell,” says Jakobi. “There was a lot less interference from the surrounding cell material than we expected.”
Having shown that it is possible to get data from crystals within a cell, the group now hopes to harness the natural ability of the peroxisome to produce crystals of other proteins, thereby side-stepping the need for laborious crystallisation experiments. “This could become a complementary method for structural biologists studying challenging proteins,” Wilmanns concludes.