Researchers identify potential new leukemia drug target
New treatment options are badly needed for acute myeloid leukemia, a relatively rare form of cancer. The malignancy begins in the bone marrow, and from there can spread rapidly to the bloodstream, depriving the body of the essential blood cells that carry oxygen and fight infections.
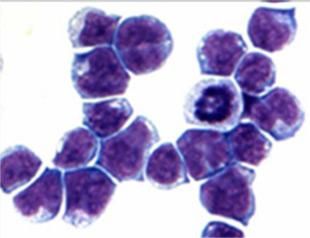
The image shows cancerous mouse bone marrow cells generated by the mutant protein AE, found in 15 percent of acute myeloid leukemia patients. Using AE as an entry point, the researchers found another protein that prompts similar changes in gene activation.
Laboratory of Biochemistry and Molecular Biology at The Rockefeller University/Nature
Now, new work from a team lead by Rockefeller University researchers has revealed a potential genetic weakness of the disease, offering insights into the molecular mechanisms behind acute myeloid leukemia and suggesting a new target for drug development.
Previously, researchers identified a variety of mutations associated with this disease, including a DNA rearrangement found in about 15 percent of patients. The abnormal DNA-binding protein produced as a result of this mutation takes on entirely new functions, dramatically altering a set of genes that are turned on in a cell to promote the cancer. But how this mutation affects these changes has remained mysterious.
In their work the researchers describe how they identified the molecular mechanism behind this gene activation.
The researchers, led by Robert G. Roeder, Arnold and Mabel Beckman Professor and head of Rockefeller's Laboratory of Biochemistry and Molecular Biology, began by searching for proteins that interact with the mutant protein, known as AE, produced by a DNA rearrangement. Their screen identified JMJD1C, an enzyme that removes chemical tags, known as methyl groups, from histones, which are proteins contained in chromosomes. These tags serve as repressive marks, indicating that genes in the associated region should be turned off.
To investigate the relationship between JMJD1C and AE, the team first explored the broader effects of removing JMJD1C. "We found that numerous genes were down-regulated upon loss of JMJD1C, and the set overlaps significantly with the genes that are normally activated by AE," explains first author Mo Chen, a postdoc in Roeder's lab.
The loss of gene expression turns out to have dramatic consequences for the disease. The team found that acute myeloid leukemia cells are addicted to the presence of JMJD1C, and without it they cannot survive. "In fact, these cells were very sensitive to depletion of JMJD1C," says Chen. "We see an increase in apoptosis, a sort of cellular suicide."
The team confirmed that JMJD1C interacts with AE, and demonstrated that the enzyme is required for AE to exert its cancer-promoting effects. But they also found that JMJD1C plays an even a broader role in acute myeloid leukemia, beyond its interaction with AE.
"We were very surprised to find that JMJD1C is required for the proliferation of other acute myeloid leukemia cell lines, which do not have AE, so we looked for other proteins that might be responsible for JMJD1C addiction," says Chen. The team found at least two other proteins that can recruit JMJD1C to target genes in diseased cells that lack AE, fueling leukemia growth.
These results suggest that JMJD1C may play a general role in promoting growth in myeloid leukemias, according to the researchers. "We are excited because this type of general phenomena is an ideal target for drug development," Roeder says.
There are already small molecules that inhibit this class of enzymes. "Our work will facilitate the development of selective inhibitors against JMJD1C, which is a highly promising therapeutic target for multiple types of leukemia," Roeder adds.























































