Blinded by genetic mutation
Team characterizes genetic mutations linked to a form of blindness
Advertisement
achromatopsia is a rare, inherited vision disorder that affects the eye's cone cells, resulting in problems with daytime vision, clarity and color perception. It often strikes people early in life, and currently there is no cure for the condition.
One of the most promising avenues for developing a cure, however, is through gene therapy, and to create those therapies requires animal models of disease that closely replicate the human condition.
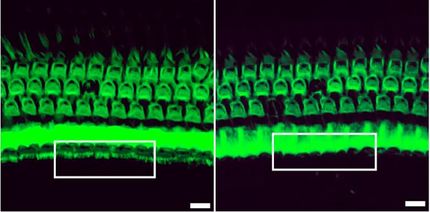
In a new study, a collaboration between University of Pennsylvania and Temple University scientists has identified two naturally occurring genetic mutations in dogs that result in achromatopsia. Having identified the mutations responsible, they used structural modeling and molecular dynamics on the Titan supercomputer at Oak Ridge National Laboratory and the Stampede supercomputer at the Texas Advanced Computing Center to simulate how the mutations would impact the resulting protein, showing that the mutations destabilized a molecular channel essential to light signal transduction.
The findings provide new insights into the molecular cause of this form of blindness and also present new opportunities for conducting preclinical assessments of curative gene therapy for achromatopsia in both dogs and humans.
"Our work in the dogs, in vitro and in silico shows us the consequences of these mutations in disrupting the function of these crucial channels," said Karina Guziewicz, senior author on the study and a senior research investigator at Penn's School of Veterinary Medicine. "Everything we found suggests that gene therapy will be the best approach to treating this disease, and we are looking forward to taking that next step."
The research began with a German shepherd that was brought to Penn Vet's Ryan Hospital. The owners were worried about its vision.
"This dog displayed a classical loss of cone vision; it could not see well in daylight but had no problem in dim light conditions," said Aguirre, professor of medical genetics and ophthalmology at Penn Vet.
The Penn Vet researchers wanted to identify the genetic cause, but the dog had none of the "usual suspects," the known gene mutations responsible for achromatopsia in dogs. To find the new mutation, the scientists looked at five key genes that play a role in phototransduction, or the process by which light signals are transmitted through the eye to the brain.
They found what they were looking for on the CNGA3 gene, which encodes a cyclic nucleotide channel and plays a key role in transducing visual signals. The change was a "missense" mutation, meaning that the mutation results in the production of a different amino acid. Meanwhile, they heard from colleague Dixon that he had examined Labrador retrievers with similar symptoms. When the Penn team performed the same genetic analysis, they found a different mutation on the same part of the same gene where the shepherd's mutation was found. Neither mutation had ever been characterized previously in dogs.
"The next step was to take this further and look at the consequences of these particular mutations," Guziewicz said.
The group had the advantage of using the Titan and Stampede supercomputers, which can simulate models of the atomic structure of proteins and thereby elucidate how the protein might function. That work revealed that both mutations disrupted the function of the channel, making it unstable.
"The computational approach allows us to model, right down to the atomic level, how small changes in protein sequence can have a major impact on signaling," said MacDermaid, assistant professor of research at Temple's Institute for Computational Molecular Science. "We can then use these insights to help us understand and refine our experimental and clinical work."
Original publication
Naoto Tanaka, Emily V. Dutrow, Keiko Miyadera, Lucie Delemotte, Christopher M. MacDermaid, Shelby L. Reinstein, William R. Crumley, Christopher J. Dixon, Margret L. Casal, Michael L. Klein, Gustavo D. Aguirre, Jacqueline C. Tanaka, Karina E. Guziewicz; "Canine CNGA3 Gene Mutations Provide Novel Insights into Human Achromatopsia-Associated Channelopathies and Treatment"; PLOSone; 2015