A paradigm shift in multidrug resistance
Advertisement
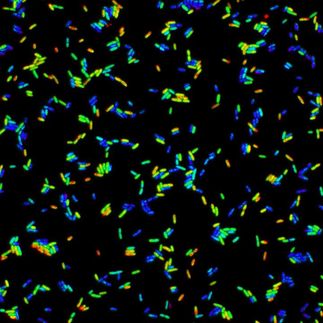
bacteria are pretty wily creatures. Take for example, an organism such as Salmonella, which are killed by antibiotics in lab tests, but can become highly resistant in the body.
Bacteria switch between antibiotic susceptible-to-resistant states during infection using a Trojan horse strategy
Peter Allen
It is an example of what UC Santa Barbara biologist Michael Mahan refers to as the Trojan horse strategy. Identified through new research conducted by Mahan and his colleagues, the Trojan horse strategy may explain why antibiotics are ineffective in some patients despite lab tests that predict otherwise. The research findings appear in EBioMedicine.
"We are not petri plates, and we need to revisit the way antibiotics are developed, tested and prescribed," said Mahan, a professor in UCSB's Department of Molecular, Cellular, and Developmental Biology. Current methods for testing resistance to antibiotics do not reflect the actual and varying environments in the body, where bacteria fight to survive. Mahan noted that this difference can render antibiotic susceptibility testing inaccurate.
"Prescription of the wrong antibiotic may not only fail to clear the infection but may create the perfect storm for the emergence of superbugs in infected patients," he added. "Even in our most advanced hospitals, high drug doses are given to infected patients without knowing that the body's environment may render bacteria inherently resistant to the very antibiotics prescribed to control them."
Mahan's research demonstrates two important points: Bacteria become resistant only to certain antibiotics, and they deploy this defense mechanism only in certain areas of the body. This means that when a patient fails to respond to a particular course of antibiotics that lab tests predict should be effective, rather than increasing the dose or duration of treatment, a potentially more effective therapeutic option is simply to prescribe another drug.
Mahan and his colleagues initially worked with the aforementioned Salmonella, a bacterium that causes food and blood poisoning. Salmonella reside within white blood cells - the very cells of the immune system that are involved in protecting the body against infectious disease. When the researchers mimicked this intracellular environment in the lab, the bacteria became highly resistant to certain antibiotics.
Next, the researchers tested Yersinia, a bacterium that also causes food and blood poisoning but lives outside of host cells in the intestine. When researchers mimicked the extracellular environment of the intestine, this bacterium also became highly resistant to certain antibiotics. These two examples indicate that the resistance process may be shared by many different types of bacteria.
"Our research suggests the need for animal models to be incorporated early during the antibiotic development process and for lab drug sensitivity testing to incorporate media that mimic the specific biochemical environments that trigger resistance in the body," Mahan explained. "If these additional components were added, we'd get a more accurate reflection of what happens when a patient is treated with a particular drug.