Clamshell-shaped protein puts the 'jump' in 'jumping genes'
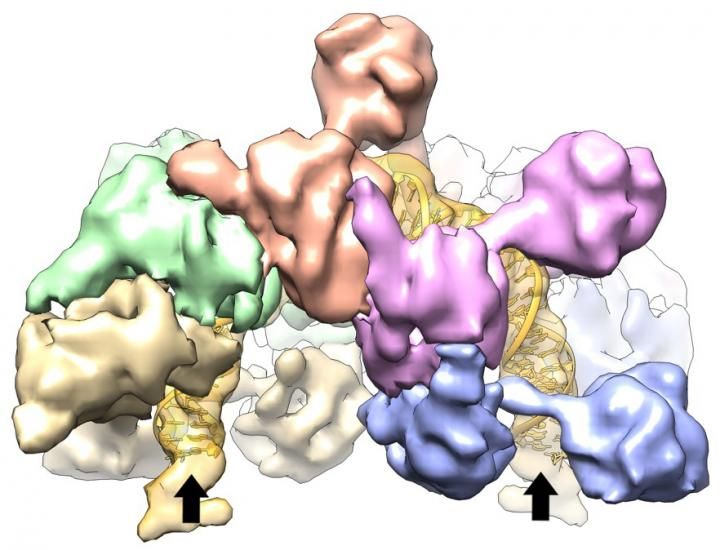
Scientists at Johns Hopkins report they have deciphered the structure and unusual shape of a bacterial protein that prepares segments of DNA for the insertion of so-called jumping genes. The clamshell shape, they say, has never before been seen in a protein but connects nicely with its function: that of bending a segment of DNA into a 180-degree U-turn.
Cell Press
The finding, they say, advances the scientific understanding of how DNA segments get moved around among bacteria, often bringing with them genes that confer toxicity or antibiotic resistance.
The protein, called IstB, is genetically similar to a whole family of related proteins found in bacteria, plants and animals, so the Johns Hopkins team says it was surprised to learn that its structure and function are not so similar to those of its family members. "What we learned is that IstB showcases the ability of natural selection to find new uses for class of enzyme that's been around a very long time," says James Berger, Ph.D., professor of biophysics and biophysical chemistry at the Johns Hopkins University School of Medicine.
At the heart of the research, Berger says, is a piece of DNA, called IS21, which is a type of transposon, or jumping gene. These segments of DNA hold the blueprint for making proteins that can cut and otherwise manipulate DNA to leave one spot in a genome and land elsewhere.
On its own, Berger points out, a jumping gene is neither helpful nor harmful but can become so if, for example, it inserts itself into and disrupts a normally functional gene. "Or it could take a nearby gene with it when it jumps," he adds. "That gene will then be in new surroundings and under new control, which could be helpful or harmful to an organism depending on the gene's function."
Berger says he and postdoctoral fellow Ernesto Arias-Palomo, Ph.D., were aware that IS21 is found in some disease-causing bacteria, like plague's Yersinia pestis, and close to genes that make those bacteria toxic to people. "That made us suspect it might play a role in moving those genes around, which can ultimately cause the transfer of harmful genes to previously harmless bacteria," he says.
To examine the idea, the team focused on IstB, one of the two proteins IS21 encodes. The other encoded protein is IstA, which cuts and pastes the IS21 transposon. IstB is its helper protein.
The team knew based on IstB's genetic sequence that it contains a "reactor" site that extracts the energy found in certain chemical molecules and uses it to alter DNA. But how it works and how it collaborates with IstA was a mystery.
Using X-rays and beams of electrons to probe IstB's structure and function, the researchers found that IstB is made of 10 smaller units, which are organized into two parallel horseshoes of five units each. Double strands of DNA fit in between.
"What we saw is that IstB can take a straight piece of DNA and bend it 180 degrees, or take a bent piece of DNA and hold it in that position," says Berger. "That primes the DNA for the insertion of a transposon."
Biochemical tests further showed that IstA recognizes the bent DNA bound by IstB and kicks off the IstB, probably to make the DNA accessible for the cuts it will make, explains Berger.
Original publication
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.
Most read news
More news from our other portals
Last viewed contents
Buttocks
A glow stick that detects cancer?
Innovative device allows 3-D imaging of the breast with less radiation
Claude_Bourgelat
Rumination_(eating_disorder)
Molecular middle managers make more decisions than bosses



















































