Method reveals what bacteria sense in their surroundings
Knowing how environmental signals modulate bacterial behavior could help combat biofouling and antibiotic resistance
A new, rapid method is helping detect how bacteria sense and respond to changes in their environment. Bacteria can pick up external signals, which then relay to internal signaling pathways that direct their behavior. This surveillance also can trigger survival tactics for a variety of harsh situations, such as lack of nutrients or the presence of antibiotics.
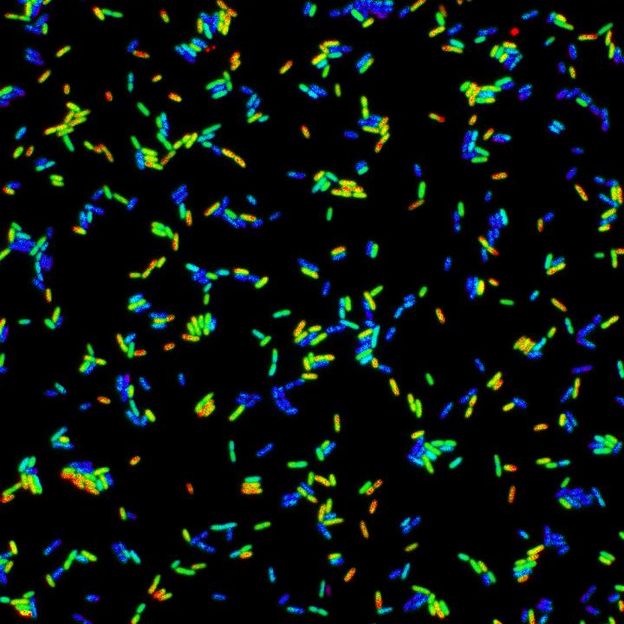
In a screening method to detect signals that regulate bacteria behavior, a biosensor is at work in Salmonella.
Erez Mills/Miller Lab/Univ. of Wash.
In an article in Science Signaling, microbiologists expressed particular interested in the sensing done by the food-poisoning bacteria, Salmonella. The pathogen is adept at adjusting to different locations and surrounding conditions. One of its safety strategies is to gather into a biofilm: a collection of bacteria huddled together in a protective coating.
Salmonella biofilms can grow persistently on many surfaces and can make the pathogen resistant to infection control measures. This is significant problem in the food-processing industry. According to the Centers for Disease Control and Prevention, Salmonella causes more than 1.2 million episodes of illness a year in the United States alone.
To adapt so readily, noted the authors of the report, Salmonella typhimurium bacteria need to figure out if they are in the stomach, within cells, or on a plant or other surface. Salmonella typhimurium is the most studied bacterial pathogen; yet, the researchers added, little is known about what Salmonella typhimurium senses in the environment.
The researchers were Erez Mills, Erik Petersen, and Bridget R. Kulasekara, working in the University of Washington laboratory of Samuel I. Miller, professor of medicine, microbiology, immunology, and genome sciences.
They describe a new screen they designed to identify the environmental cues Salmonella picks up and that, in turn, modulate the production of a second messenger molecule.
This molecule, cyclic-di-GMP, governs bacterial motility and biofilm formation in many species of bacteria in response to environmental conditions, but up to now specific signals that modulate cyclic-di-GMP in Salmonella were largely unknown.
The researchers discovered that many environmental signals modulate cyclic-di-GMP in Salmonella typhimurium and subsequently altered its biofilm formation.
Their screen for these signals and their actions was based on a combination of leading-edge technologies, including a genetically engineered biosensor for cyclic-di-GMP, used with either microscopy or flow cytometry, a laser-based measurement on single cells suspended in a fluid.
"The most interesting signal we identified was L-arginine, one of the twenty commonly used amino acids," the researchers noted. They explained that Salmonella typhimurium specifically responds to very low concentrations of L-arginine, in amounts too little to be utilized as a nutrient source.
The researchers think the sensing of these low levels of L-arginine tells the bacterium that it is in a certain type of environment and prompts the bacterium to adapt accordingly.
Future work, they said, will be aimed at determining the types of environments where L-arginine is sensed, as well as the bacterium's behavior in response to L-arginine.
In addition to providing new information on how Salmonella typhimurium senses chemical signals in its environment, the method the scientists designed can be used in studies of other bacterial species to increase knowledge about microbial sensing.
Possible applications range from combating antibiotic resistant bacteria to ameliorating agricultural and industrial biofouling.