New degradation proteins show route to cell survival
Studies by researchers at Tokyo Institute of Technology and colleagues reveal two proteins that induce degradation of certain cell constituents to help cell survival under nutrient-limiting conditions.
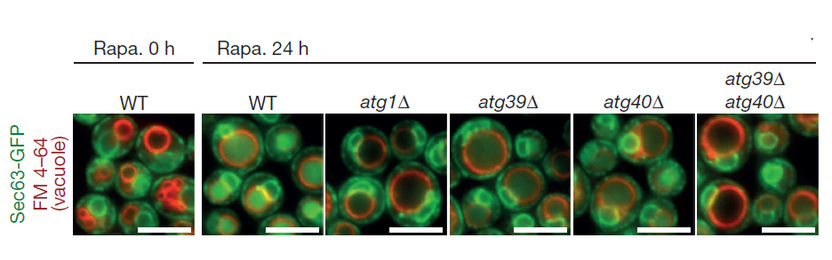
Fluorescence microscopy images of yeast cells. Left to right: untreated wild type cells (Rapa. 0 h); cells treated with rapamycin for 24 hours (Rapa. 24 h) - wild type, atg1 mutant, atg39 mutant, atg40 mutant and atg39 atg40 double mutant. The researchers used the ER membrane protein Sec63 fused with green fluorescent protein (GFP) to monitor autophagy of the ER. Autophagic degradation of Sec63–GFP yielded vacuolar protease-resistant GFP fragments, which increased GFP fluorescence within the vacuole (red circles).
The degradation of cell constituents helps maintain the functions of the cell. This degradation process – ‘autophagy’ – is linked to several diseases. Although recent studies have suggested that selective autophagy targeted at specific cell constituents may also be responsible for maintaining healthy conditions in the cell – ‘homeostasis’ – the extent of these processes is unknown. Now researchers at Tokyo Institute of Technology, Yokohama City University and the Japan Science and Technology Agency have identified two new proteins that drive selective autophagy. They also show the role of these new autophagy pathways in preserving the cell in nutrient-limiting conditions.
The researchers focused their study on the yeast Saccharomyces cerevisiae. During selective autophagy, an autophagosomal membrane extends over the target through the binding of autophagy receptors to Atg8 proteins. By looking for proteins that bound to Atg8 in the yeast, Hitoshi Nakatogawa and colleagues identified two new autophagy receptor proteins: Atg39 and Atg40.
The researchers noted that Atg39 and Atg40 levels increased in the presence of the chemical rapamycin, which mimics nitrogen starvation. Further studies linked the two proteins to autophagy of a certain cell constituent – the endoplasmic reticulum (ER), a network of flattened membrane enclosed sacks – in nitrogen-starved conditions. The same conditions also triggered degradation of a part of the nucleus by Atg39; this protein localized to a special part of the ER surrounding the nucleus. Atg40 localized to other ER regions and mediated their degradation. Blocking nucleus degradation by Atg39 led to cell death in nitrogen-starved conditions. Atg40 was suggested to correspond to a human protein encoded by FAM134B, a causative gene for a hereditary sensory and autonomic neuropathy.
“Our results provide fundamental insight into the pathophysiological roles and mechanisms of ‘ER-phagy’ and ‘nucleophagy’ in other organisms,” explain the researchers in their report of the work. Further studies are required to determine the ER components that must be degraded as well as whether the purpose of this autophagy process is removal of material or making substances available through the breakdown.
Original publication
Original publication
Keisuke Mochida, Yu Oikawa, Yayoi Kimura, Hiromi Kirisako, Hisashi Hirano, Yoshinori Ohsumi & Hitoshi Nakatogawa; "Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus."; Nature.
Organizations
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.





















































