A molecule in an optical whispering gallery
With the help of a microsphere and a nanowire, single unlabelled biomolecules can be detected through light
Being able to track individual biomolecules and observe them at work is every biochemist’s dream. This would enable the scientists to research in detail and better understand the workings of the nanomachines of life, such as ribosomes and DNA polymerases. Researchers at the Max Planck Institute for the Science of Light have taken a big step closer to this goal. Using an optical microstructure and gold nanoparticles, they have amplified the interaction of light with DNA to the extent that they can now track interactions between individual DNA molecule segments. In doing so, they have approached the limits of what is physically possible. Their optical biosensor for single unlabelled molecules could also be a breakthrough in the development of biochips: fingernail-size mini-labs in mobile analytical devices could test a drop of blood for multiple diseases simultaneously or facilitate comprehensive environmental analyses with very little sample material.

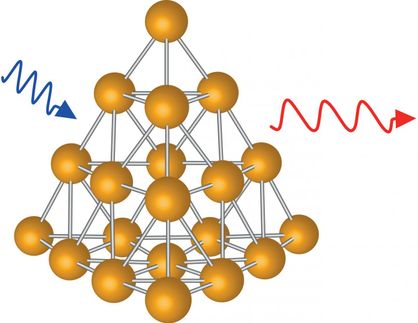
A team of Max Planck researchers headed by Frank Vollmer attach gold wires to microspheres, to which molecules are able to bind. The molecules can be detected because the wavelength of the light shifts and is amplified by the microsphere and nanowire.
© MPI for the Science of Light
Our understanding of fundamental life processes was first made possible by knowledge of how individual biomolecules interact with each other. In cells, nanomachines such as ribosomes and DNA polymerases stitch individual molecules together to form complex biological structures such as proteins and DNA molecules, the repositories of genetic information. Although it is possible to investigate the interaction of individual molecules with enzymes or ribosomes, the molecules often have to be labelled, for example with fluorescent markers, in order to observe them. However, such labelling is only possible with certain molecules, and it can interfere with the function of the biological nanomachines. Although light can be used to detect unlabelled biomolecules, the approach cannot be used to detect single DNA molecules, as the interaction of light waves with the molecule is too weak.
A team of physicists headed by Frank Vollmer of the Laboratory for Nanophotonics and Biosensors at the Max Planck Institute for the Science of Light has now succeeded in amplifying the interaction of light with DNA molecules to the extent that their photonic biosensor can be used to observe single unlabelled molecules and their interactions.
A microsphere becomes an optical whispering gallery
To achieve this, the physicists use glass beads around 60 micrometres in diameter, about the thickness of a human hair, and gold nanowires approx. 12 nanometres in diameter and 42 nanometres in length. The gold wire is therefore only about one ten-thousandth the thickness of a hair. The microsphere and nanowire amplify the interaction between light and molecules. With the help of a prism, the researchers shine laser light into the microsphere. The light is repeatedly reflected at the internal surface of the sphere until, ultimately, it propagates along the inside surface, similar to the way sound waves travel along the walls of a circular enclosure or whispering gallery: when someone whispers at one end of the domed or vaulted gallery, a person at the opposite end can hear it on the other end, even over an unusually long distance. This is because the sound waves do not lose intensity as they travel.
If a molecule is fixed to the surface of the glass bead, the light beam travels past it more than a hundred thousand times. Because the light wave always extends somewhat outside the microsphere, an interaction occurs between it and the molecule. This interaction is greatly amplified due to the frequent contact between the light and the molecule. However, the interaction is still too weak to register single molecules.
Vollmer and his colleagues therefore fix a nanowire to the surface of the glass bead. The light whizzing past generates plasmons: collective oscillations of electrons. “The plasmons pull the light wave a little further out of the glass microsphere,” Vollmer explains. This amplifies the field strength of the light wave by a factor of more than a thousand. The gain in signal is then sufficient to detect single biomolecules, such as DNA fragments. The Erlangen-based researchers did precisely that. They attached a fragment of single-strand DNA, which always occurs in the form of a double strand in the cell nucleus, to the nanowire mounted on the microsphere. When a matching, i.e. complementary, DNA fragment binds to the “bait” on the nanowire, the wavelength of the light shifts and is amplified by the microsphere and nanowire. This shift can be measured.
Different strand sections can be distinguished by their binding behaviour
However, the physicists used a shorter DNA fragment than is usual in similar procedures. Like a short piece of tape on a wall, short DNA fragments do not adhere strongly to each other, so that the strands separate again relatively quickly. Hence, new fragments are able to bind repeatedly to the molecular “bait”, including fragments that are not fully complementary. In this way, it is possible to investigate how long the DNA fragments interact with each other and how often the “bait” captures a segment. “This approach makes it possible to use a single DNA receptor and to follow its successive interactions with various DNA segments in the sample solution,” says Frank Vollmer. “Based on the duration and frequency of the measured interactions, it is then possible to detect specific unlabelled DNA molecules.”
The researchers have tested their optical biosensor with a sample containing both an exactly matching DNA fragment and a fragment that was not perfectly complementary. They were able to distinguish the two fragments based on their different kinetics.
Even in nature, the bonds formed between molecules and nanomachines are fleeting. Thanks to the new method, it is now possible to explore such natural kinetics in greater detail, says Frank Vollmer. “More research is needed,” says the physicist, who is looking forward to tackling future challenges.
The researchers in Erlangen are already planning future projects. “It’s possible to observe, for example, how an enzyme such as DNA polymerase synthesizes DNA ,” explains Vollmer. The scientists would also like to integrate their photonic biodetector into optical microchips for use in clinical diagnostics.