Conserved nuclear envelope protein uses a shuttle service
Researchers at the Stowers Institute for Medical Research have glimpsed two proteins working together inside living cells to facilitate communication between the cell's nucleus and its exterior compartment, the cytoplasm. The research provides new clues into how a crucial protein that is found in organisms from yeast to humans does its work.
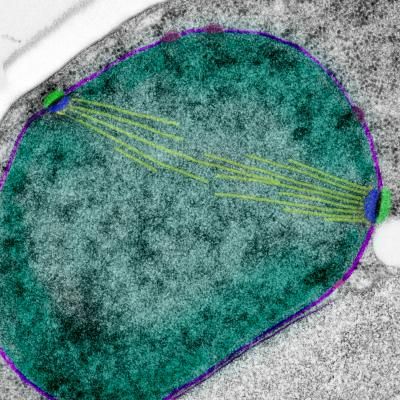
The conserved nuclear protein Ndc1 controls the insertion of holes into the nuclear membrane (shown in bluish purple). These openings become the sites for two essential structures: passageways called nuclear pore complexes (shown in purple), and spindle pole bodies (shown in green and blue), which anchor the microtubules (shown in yellow) that pull chromosomes to opposite sides of a dividing cell. The nucleoplasm is shown in dark green and the cytoplasm in grey.
Courtesy of Christine Smoyer, Stowers Institute for Medical Research
The study, led by Stowers Investigator Sue Jaspersen, Ph.D., focused on a protein called Ndc1, which controls when and where a cell inserts holes into the double-walled membrane that surrounds its nucleus. In yeast, these holes become the sites for two essential structures: passageways called nuclear pore complexes, and spindle pole bodies, which anchor the cytoskeletal filaments that pull chromosomes to opposite sides of a dividing cell.
"Too many or too few insertion sites will have disastrous consequences," Jaspersen says, explaining that new nuclear pore complexes and spindle pole bodies must be created each time a cell prepares to divide to ensure genetic material can be properly distributed and daughter cells are equipped for gene activation and protein production. Her team's findings on Ndc1's interactions with a protein called Mps3, which appears to govern Ndc1's distribution on the nuclear envelope, are described in the issue of the Journal of Cell Biology.
Jaspersen and her colleagues set out to study Ndc1 because it is absolutely crucial for cell survival. They knew that in yeast, Ndc1 is embedded in the nuclear envelope and is needed for the insertion of both nuclear pore complexes and spindle pole bodies. But because cells are so sensitive to changes in Ndc1, scientists had been unable to learn much about how the protein functions.
Traditional genetic strategies of eliminating, altering, or increasing Ndc1 to test its function typically killed cells. "Having the exact right amount of Ndc1 is really critical," Jaspersen says. "That makes working with the gene very technically challenging. So we knew it was really important and the amount was really important, but we didn't know what it did or why it did it."
To learn more, Jaspersen and Jingjing Chen, a postdoctoral researcher in her lab, created yeast with mutations in the ndc1 gene. As expected, changes that disrupted Ndc1's interaction with its known partners in the nuclear core complex or the spindle pole body were lethal. One mutation puzzled the scientists, however. The altered Ndc1 protein bound to the expected components of both the nuclear pore complex and the spindle pole body, just like the normal protein. "Previous data suggests that if Ndc1 can functionally bind to these components, both the spindle pole body and the nuclear pore complex should be fine," Chen says. But when yeast cells produced this altered version of Ndc1, they died. "So this suggests that there may be another very critical interaction partner," she says.
Chen devised a strategy to search for such a partner. She turned to a method called a yeast two-hybrid assay, in which researchers link the proteins they are testing to two different parts of a gene activator. If the proteins associate with one another inside the cell, they bring along the gene activator components, allowing them to work together to switch on an easily detectable gene. Using a membrane-based yeast two-hybrid assay, which could detect interactions at the nuclear envelope, Chen found that Ndc1 bound to a protein called Mps3.
Jaspersen had first encountered Mps3 in 2002, when she discovered that yeast needed the protein to duplicate their spindle pole bodies prior to cell division. Mps3's interaction with Ndc1, however, was a surprise.
The institute's molecular microscopy research advisors, Brian Slaughter, Ph.D., and Jay Unruh, Ph.D., proposed that the team use a sophisticated imaging technique called fluorescence cross-correlation microscopy to look directly at the Mps3 protein inside living cells and watch for its interactions with Ndc1. To monitor Mps3's interaction with Ndc1, the researchers attached a fluorescent red tag to Ndc1 and a fluorescent green for Mps3. They then focused their microscope on a specific point within a living cell, and watched for green- or red-tagged proteins to enter or leave that space. Their observations would reveal whether Ndc1 and Mps3 were moving independently within the cell, or whether they were physically linked.
"This allowed us to look inside living yeast cells to study the complexes in real time, in the context of the nuclear envelope. We could just sit there and watch, rather than touching or breaking open the cells," Jaspersen says. The imaging not only confirmed the interaction between Mps3 and Ndc1, it allowed the team to track exactly where it occurred.
Although she and her colleagues expected to see Mps3 associate with Ndc1 either at nuclear pore complexes or at the cell's spindle pole bodies, neither turned out to be true. Instead, Mps3 and Ndc1 came together in the nuclear envelope, but away from the two structures the team had set out to study. This led them to speculate that Mps3 might help shuttle Ndc1 to the sites where it is needed, controlling the distribution of nuclear pore complexes and spindle pole bodies. Jaspersen team is now planning further experiments to test how Ndc1 and Mps3 maintain the appropriate balance of these critical structures.