Making and breaking heterochromatin
Scientists discover 2 further mechanisms necessary for the formation of heterochromatin
To fit the two-meter long DNA molecule into a cell nucleus that is only a few thousandths of a millimetre in size, long sections of the DNA must be strongly compacted. Epigenetic marks maintain these sections, known as heterochromatin. Scientists of the Max Planck Institute of Immunobiology and epigenetics in Freiburg have now discovered two further mechanisms necessary for the formation of heterochromatin. The research group, led by Thomas Jenuwein, describes two novel enzymes, Prdm3 and Prdm16, which attach a methyl group to a particular packaging protein of the DNA. These epigenetic marks assure that heterochromatin, and with it the structure of the cell nucleus, remain intact. Moreover, in an additional study they have determined that transcription factors bind within heterochromatin and repress the output of non-coding RNA. In contrast to less densely compacted regions known as euchromatin, in which the transcription factors accumulate at specific sites, the binding sites of transcription factors in heterochromatin are much more randomly distributed.
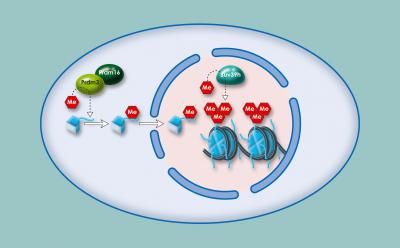
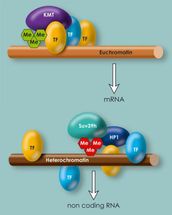
Methylation of histones within heterochromatin: The enzymes Prdm3 and Prdm16 attach a methyl group to histone H3. In the cell nucleus, Suv39h attaches two further methyl groups. The cell appears to require this triple methylation for heterochromatin to be stable.
Art For Science
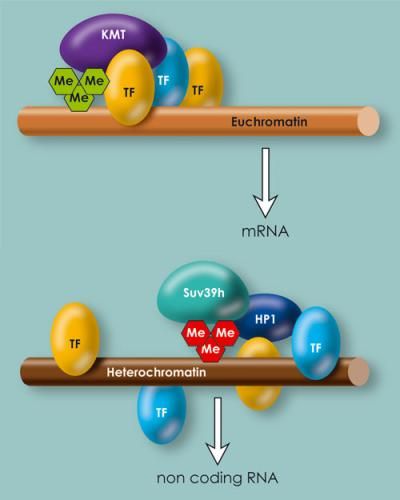
In euchromatin and heterochromatin, the binding locations for transcription factors are distributed differently. In euchromatin they are concentrated at specific locations, in heterochromatin they are distributed more randomly.
Art For Science
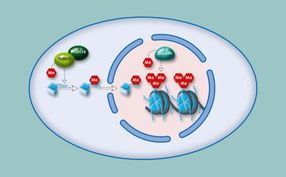
Chromatin consists of the DNA molecule and numerous proteins, including histones, which act as packaging proteins. In contrast to the easily accessible euchromatin, which contains the majority of genes, the densely compacted heterochromatin is mostly made of up of repetitive sequences that are able to form non-coding RNA-molecules.
Heterochromatic sections are found at centromeres and at chromosome ends, the telomeres. Chemical modifications of histones can alter the degree to which chromatin is compacted. For example, methyltransferases add methyl groups to proteins at various positions. These epigenetic alterations regulate the formation and maintenance of heterochromatin.
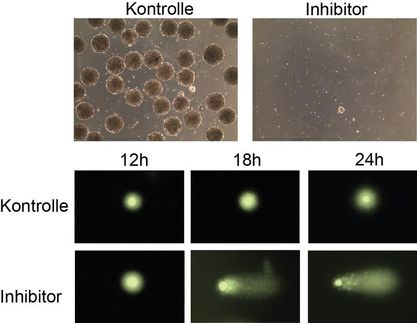
Inês Pinheiro, a doctoral student in Thomas Jenuwein's department, has now discovered that Prdm3 and Prdm16 function as methyltransferases and attach a methyl group to histone H3 at the lysine 9 (H3K9) position. Until now, both proteins were thought to be just transcription factors, regulating the activity of various genes. Experiments in which the Freiburg-based researchers switch off both enzymes demonstrate how important Prdm3 and Prdm16 are. Heterochromatin breaks down and the heterochromatin regions can be read. "Our experiments show that Prdm3 and Prdm16 attach a methyl group at H3K9. This single-methylated H3 (H3K9me1) is then transported into the cell nucleus and inserted into heterochromatin. Only then heterochromatin remains intact," explains Thomas Jenuwein, Director at the Max Planck Institute of Immunobiology and Epigenetics. Other methyltransferases, such as Suv39h, can add another two methyl groups (H3K9me3) to the single-methylated histone and thus further increase heterochromatin stability.
Moreover, the researchers in Freiburg observed that the lamina of the cell nucleus is impaired without Prdm3 and Prdm16. Heterochromatin must be associated with this layer of lamina proteins at the inner nuclear membrane. "The cell apparently requires methylation at H3K9 and a yet-unknown chromatin or lamina protein by Prdm3 and Prdm16 for heterochromatin to be stable. As with other methyltransferases, we assume that both enzymes can methylate other molecules besides histones. However, we do not know whether the destruction of the lamina is triggered by the loss of heterochromatin or by the absence of methylation at a lamina protein," says Jenuwein.
However, it is not just methylation of histones that is necessary to maintain heterochromatic regions. In a further study, doctoral students Aydan Karslioglu and Valentina Perrera examined the role of transcription factors, i.e. proteins that bind to DNA and control gene activity – in the case of heterochromatin, the repression of non-coding RNA molecules. This study showed that two transcription factors are essential for intact heterochromatin: Pax3 and Pax9. Only when both factors and their binding sites are present in the repetitive DNA heterochromatin remains intact. The researchers assume, however, that additional transcription factors can also bind to repetitive sequences in heterochromatin.
Transcription factors thus control gene activity in euchromatin, as well as in heterochromatin. Despite this, there are differences between the two. In heterochromatin, the binding locations are distributed comparatively at random over the DNA strand, whereas euchromatin is concentrated at the locations important for gene regulation. "In our data, the distribution within heterochromatin looks like the Aigulles Droites in the Mont Blanc massif: a lot of small peaks without deep valleys in between. Euchromatin looks more like the Matterhorn: one high peak without secondary peaks," as Thomas Jenuwein describes the results.
For the researchers, an important difference between heterochromatin and euchromatin lies in the control of gene activity and the formation of RNA. "In heterochromatin, the binding sites for transcription factors are distributed more randomly, so that they cannot reinforce or intensify one another's effect. The DNA therefore cannot be read in such a precise and coordinated manner at these locations. Inhibiting influences that largely turn off heterochromatin dominate in the end," says Jenuwein. With euchromatin, in contrast, the transcription factors bind to DNA in such a way that they enhance each other's function. This permits precise control over the gene activity.
Original publication
I Pinheiro, R Margueron, N Shukeir, M Eisold, C Fritzsch, F M Richter, G Mittler, C Genoud, S Goyama, M Kurokawa, J Son, D Reinberg, M Lachner & T Jenuwein; "Prdm3 and Prdm16 are H3K9me1 Methyltransferases Required for Mammalian Heterochromatin Integrity"; Cell 2012 (Vol. 150, Issue 5, pp. 948-960)
A Bulut-Karslioglu, V Perrera, M Scaranaro, I A de la Rosa-Velazquez, S van de Nobelen, N Shukeir, J Popow, B Gerle, S Opravil, M Pagani, et al.; "A transcription factor–based mechanism for mouse heterochromatin formation"; Nature Structural & Molecular Biology (2012)