On the trail of the epigenetic code
Test system on Drosophila should provide the key to histone function
The genetic inherited material DNA was long viewed as the sole bearer of hereditary information. The function of its packaging proteins, the histones, was believed to be exclusively structural. Additional genetic information can be stored, however, and passed on to subsequent generations through chemical changes in the DNA or histones. Scientists from the Max Planck Institute for Biophysical chemistry in Göttingen have succeeded in creating an experimental system for testing the function of such chemical histone modifications and their influence on the organism. Chemical modifications to the histones may constitute an "epigenetic histone code" that complements the genetic code and decides whether the information from certain regions of the DNA is used or suppressed.
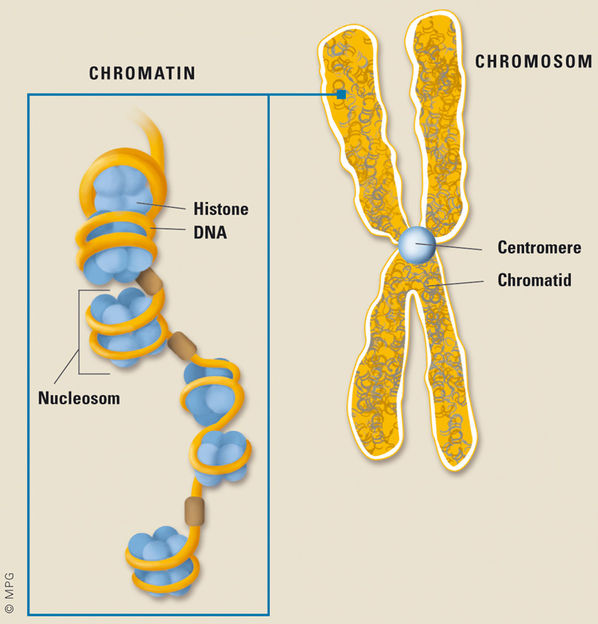
The condensation of the DNA involves a dramatic restructuring of the two metre-long DNA thread to a chromosome that has a diameter of 1.5 micrometres. The DNA is wound around the packaging proteins called "histones".
Max-Planck-Gesellschaft
How do you get a two-metre-long DNA thread into the cell nucleus? By winding it into a ball, of course! The DNA is wound around proteins known as histones, becoming 50,000 times shorter as a result. Other proteins then aggregate on it to form chromatin and, finally, the chromosomes. The latter are the product of an ingenious packaging trick. The five types of histones (H1, H2A, H2B, H3 und H4) fulfil even more tasks, however, and this is what makes them so fascinating. Histones can have small chemical attachments, such as acetyl, methyl and phosphate groups, in different places. These cause the opening of the chromatin, for example, and hence enable the genetic information to be read. Conversely, certain areas of the DNA molecule can be deactivated and rendered unreadable through other modifications, such as the binding of proteins. Scientists refer to this process as "gene silencing". "The histone modifications can intervene in the control of gene activity in this way and, as a result, complement the genetic code," explains Herbert Jäckle, Director of the Max Planck Institute for Biophysical Chemistry in Göttingen.
Every time a cell divides, this modification pattern of the histones is inherited by the daughter cells. The scientists assume that this epigenetic inheritance is controlled by a cell-specific or organ-specific "histone code". "This decides whether the cell machinery has access to the DNA-coded genes or whether the access is blocked," says Jäckle. The scientists would very much like to crack this code: for the production of the histones, hundreds of gene copies are stored in the genome of higher organisms. Therefore, up until now, it appeared to be impossible to switch off these gene copies and replace them with genetically-modified histone variants. Researchers could only create a test system if they managed to do this: if these variants lack certain docking sites, for example for chemical groups, certain modifications to the histones could be prevented and it would then be possible to investigate the extent to which the absence of these modifications leads to diagnosable defects in the organism.
The Max Planck researchers in Göttingen have now succeeded in developing a new method for researching the function of all histone modifications. The cell biologists removed all of the histone genes from the genome of the fruit fly Drosophila melanogaster. As a result, the cells could no longer divide. As occurs with normal cell division, the organism’s genome is still doubled through DNA synthesis but the cell then remains at a standstill in the division cycle and the organism dies. The situation normalises progressively, however, with the increasing number of copies of the four histone genes produced: "Flies with twelve copies of the histone gene cluster ultimately survive and are capable of reproducing," explains Jäckle’s colleague and project leader Alf Herzig.
It had already been established for multicellular organisms that several copies of the histone gene are required for the organism to survive. However, the results obtained by the researchers also indicate that the cell realises during division that histones are lacking, and the division of the cell is then halted despite the fact that DNA has already been doubled - as is the case during all cell division processes. "Communication paths clearly exist between the histone production process and the cell division machinery," says Ufuk Günesdogan, a doctoral student in the department. Most importantly, the researchers now have a test system at their disposal into which histone variants can be channelled for the gradual experimental examination of the function of histone modification and, ultimately, the histone code in the organism. It can only be a matter of time now until the code is finally cracked.
Original publication: Ufuk Günesdogan, Herbert Jäckle & Alf Herzig; "A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes"; EMBO reports (2010) 11, 772 - 776, October 1, 2010