Molecular 3D structure of viral "copying machine" deciphered
Approach for drug development
Researchers at the University Medical Center Göttingen (UMG) and the Max Planck Institute (MPI) for Multidisciplinary Sciences have shown, for the first time, how the genetic material of the Nipah virus replicates in infected cells. The virus can cause fatal encephalitis in humans. Using cryo-electron microscopy, the team led by Prof. Dr. Hauke Hillen was able to visualize the three-dimensional structure of the viral "copying machine". These findings could contribute to the future development of antiviral drugs for the treatment of Nipah virus infections. The results of the study have now been published in the journal Nature Communications.

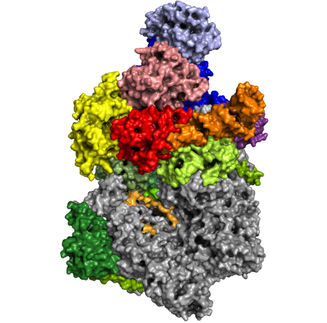
Artistic representation of the 3D structure of the RNA polymerase of the Nipah virus in the active state. The structure of the Nipah virus RNA polymerase is shown as a transparent surface representation (L protein in green, P protein in orange). The viral RNA, which serves as a template for the RNA polymerase, is shown in blue, the newly produced product RNA in red. The nucleotide substrate is shown in yellow.
umg/fernanda sala
Disease outbreaks and regional epidemics caused by viruses transmitted from animals to humans continue to occur around the world. Many pandemics that spread across national borders also originate from this mode of transmission. Early research into pathogens is essential to ensure that effective drugs and vaccines are available in the event of an epidemic or pandemic.
The World Health Organization (WHO) classifies Nipah virus as a potentially very dangerous virus for humans. It has caused several outbreaks in Asia in recent years. The virus can be transmitted from bats to humans, causing severe disease that can be fatal in up to 70 percent of cases. It can also be transmitted from person to person, spreading very quickly. There are currently no targeted drugs or vaccines available to treat Nipah virus infection.
Approach for drug development
Researchers led by Prof. Dr. Hauke Hillen, head of the "Structure and Function of Molecular Machines" research group at the Department of Cellular Biochemistry at the University Medical Center Göttingen (UMG) and research group leader at the MPI for Multidisciplinary Sciences, have succeeded in visualizing the three-dimensional structure of the Nipah virus copy machine, also known as RNA polymerase, at molecular resolution for the first time. The RNA polymerase is responsible for the replication of viral genetic material and the activation of viral genes, and is essential for the replication of the virus in cells. It is therefore a promising target for drug development. The scientists used cryo-electron microscopy to decipher the three-dimensional (3D) structure of the RNA polymerase. They shock-froze the RNA polymerase in two different states, free and bound to viral RNA, and then took thousands of individual images of the molecule in a state-of-the-art electron microscope. High-performance computers were then used to calculate a 3D structure with almost atomic resolution.
"This is an important milestone because until now it was not known exactly what the RNA polymerase of the Nipah virus looks like and how it interacts with the viral RNA. Our data show that it is similar to the RNA polymerases of other related RNA viruses, such as Ebola, but has some special features," says Prof. Hillen. The data also reveals how such a viral RNA polymerase uses the genomic viral RNA as a template for the copying process, and how it binds the newly produced product RNA and nucleotide building blocks. "These results are particularly exciting because a molecular snapshot of the RNA polymerase in its active state has never been visualized before, even for related viruses such as Ebola," says Dr Fernanda Sala, postdoctoral researcher in the research group "Structure and Function of Molecular Machines" and first author of the study. "By comparing the snapshots of free and RNA-bound RNA polymerase, we were not only able to decipher its structure, but also to gain new insights into its dynamics. Such data can be useful in the targeted development of drugs that could inhibit the RNA polymerase," she continues.