Refining the CRISPR scissors
Research team unveils novel anti-CRISPR protein mechanism for precise control of genome editing technologies
The CRISPR-Cas gene scissors offer a wide range of potential applications, from the treatment of genetic diseases to antiviral therapies and diagnostics. However, to safely harness their powers, scientists are searching for mechanisms that can regulate or inhibit the systems’ activity. Enter the anti-CRISPR protein AcrVIB1, a promising inhibitor whose exact function has remained a mystery—until now. A research team from the Helmholtz Institute for RNA-based Infection Research (HIRI) in Würzburg, in collaboration with the Helmholtz Centre for Infection Research (HZI) in Braunschweig, has uncovered the precise way AcrVIB1 works that expands the known means by which Acrs can shut down CRISPR. The results were published in the journal Molecular Cell.
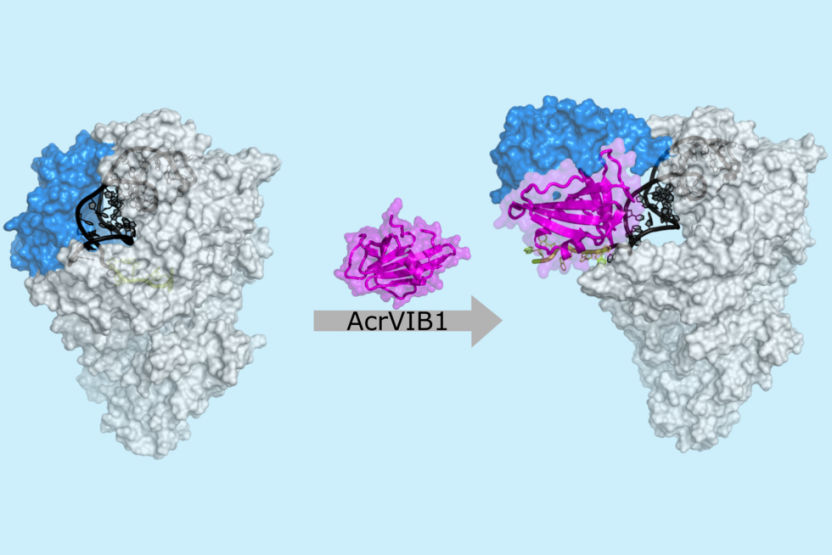
Cryo-electron microscopy image: The anti-CRISPR protein AcrVIB1 (magenta) attaches to the nuclease Cas13b (light gray) and strengthens its binding to the CRISPR RNA (crRNA; dark gray and green). AcrVIB1 induces a structural rearrangement in the nuclease (blue) and exposes the CRISPR RNA, which can then be cleaved and subsequently degraded.
© HZI / Stefan Schmelz
Bacteria and their viruses, known as phages, are locked in an age-old arms race. To defend against phage attacks, bacteria have evolved sophisticated mechanisms to recognize and counteract invading viruses. In turn, phages have developed innovative strategies to evade these defenses. A prime example of this ongoing battle is the CRISPR-Cas defense system in bacteria, countered by anti-CRISPR proteins (Acrs) in phages, which specifically block these bacterial “gene scissors”.
Apart from their counter-defensive function, anti-CRISPR proteins hold great promise for enabling more precise control over CRISPR technologies. To unlock their full potential, it is essential to understand the underlying mechanisms. Researchers at the Helmholtz Institute for RNA-based Infection Research (HIRI), a site of the Braunschweig Helmholtz Centre for Infection Research (HZI) in cooperation with the Julius-Maximilians-Universität Würzburg (JMU), and HZI scientists have now further elucidated the function of an important yet so far uncharacterized anti-CRISPR protein.
“In a previous study, we used a deep learning algorithm to predict new Acrs. This led to the identification of AcrVIB1, the first anti-CRISPR protein targeting the Cas13b nuclease,” says HIRI department head Prof. Chase Beisel, who led the study together with the department of Prof. Wulf Blankenfeldt at HZI. “The nuclease Cas13b can recognize and cut RNA. It is currently used to silence genes, whether to study their function, clear viruses, or counteract genetic diseases linked to the gene.” However, how the protein AcrVIB1 inhibits Cas13b remained unknown until now. In a study published today in the journal Molecular Cell, the research team presents this entirely new blocking mechanism.
An RNA dead end
The Cas13b nuclease operates by interacting with a CRISPR ribonucleic acid (crRNA), which serves as a guide to identify and bind to complementary RNA sequences, for example those from phages. Once the target RNA is bound, Cas13b can cleave and degrade not only these complementary RNA molecules but also all other RNAs in the vicinity. While most known anti-CRISPR proteins block steps along this path such as crRNA binding or target recognition, AcrVIB1 adopts a radically different strategy: Rather than blocking the binding of the crRNA to Cas13b, AcrVIB1 even improves it. The formed pair is dysfunctional though, meaning that the enzyme cannot begin degrading RNAs even when its target is present. Furthermore, the bound crRNA becomes vulnerable to attack by cellular ribonucleases, which break down RNA molecules.
“The tighter binding between nuclease and guide RNA was entirely unexpected. The simpler and therefore initially expected mechanism would have been to just prevent the guide RNA from binding in the first place,” says first author Dr Katharina Wandera, who completed her doctorate in Chase Beisel's laboratory. “Nevertheless, the path taken by AcrVIB1 appears to be more effective: AcrVIB1 binds tightly to and thereby renders Cas13b inactive. At the same time, it increases the turnover of guide RNAs, making Cas13b a dead end for crRNAs.”
Chase Beisel's team at HIRI and the laboratory of Wulf Blankenfeldt at HZI have joined forces to decipher the structure of the inhibition mechanism more precisely. Using cryo-electron microscopy, Blankenfeldt's group showed that AcrVIB1 binds to Cas13b, leaving the crRNA-binding domain untethered. “Our finding provides a blueprint for the development of molecules that could mimic or modify the function of the anti-CRISPR protein,” says Blankenfeldt. These are the first data from the HZI's new cryo-electron microscopy facility to be published.
A vast field
“In the future, we could use molecules such as AcrVIB1 to regulate or temporarily deactivate CRISPR systems across a variety of applications,” states Blankenfeldt. This discovery holds potential to further enhance the safety and precision of CRISPR-based technologies.
“Deciphering this mechanism also provides valuable insights into the co-evolution of bacteria and viruses, which are constantly trying to outsmart each other,” explains Wandera. The deeper understanding of bacterial resistance could play a pivotal role in the development of new antibiotics and expand the possibilities of synthetic biology.
In summary, this study not only contributes to a better understanding of anti-CRISPR strategies, but also paves the way for innovative therapies and diagnostic approaches in medicine. “But this is just the beginning: There are certainly many more Acrs and novel inhibitory mechanisms waiting to be discovered,” says Beisel, giving an outlook on future research projects.