What triggers the programmed cell death mechanism?
Development and combination of various microscopic and spectroscopic techniques
How can molecular structures be analysed when the resolution of the techniques available is not sufficient? Researchers from the fields of physics, chemistry and medicine at Heinrich Heine University Düsseldorf (HHU) have combined and further developed various microscopic and spectroscopic techniques in order to examine a protein arrangement in the cell membrane that is important for “programmed cell death”. In the scientific journal Science Advances, they now describe the circumstances under which the CD95 receptor – which is responsible for cell death – reacts.
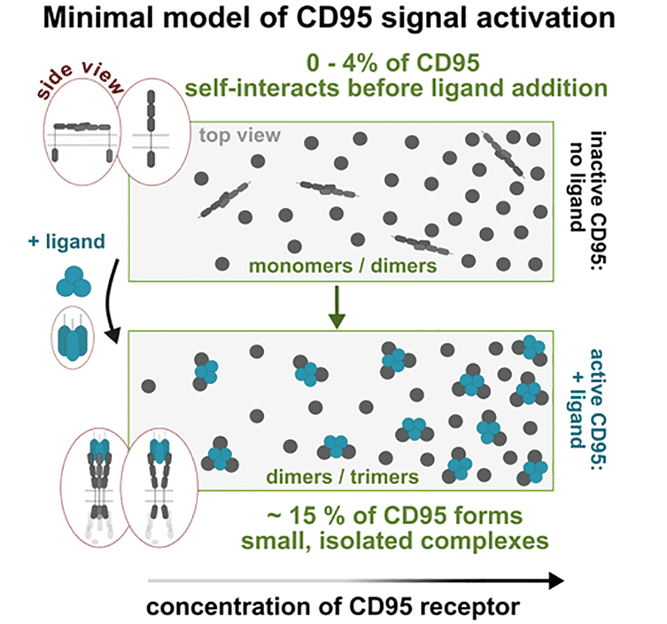
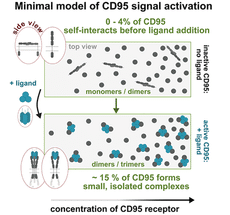
Model of the signal activation resulting from the measurements: Following interaction with the CD95 ligand, the CD95 receptor alters its arrangement and conformation, causing small protein complexes to form.
HHU / Cornelia Monzel
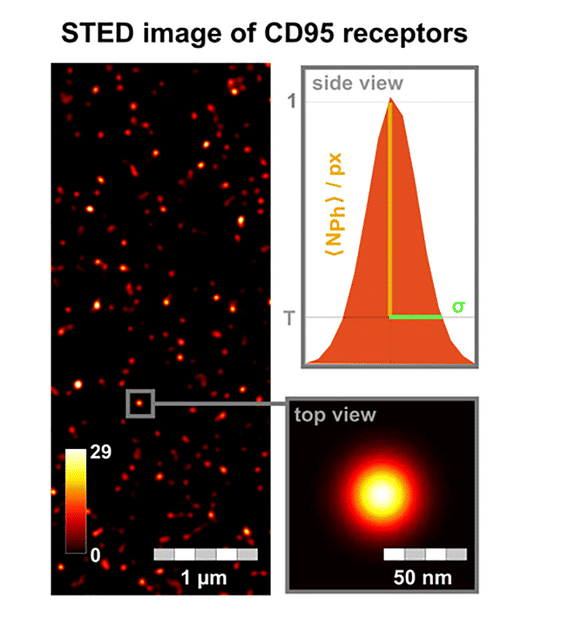
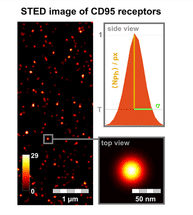
STED image of CD95 receptors on the cell membrane. For every fluorescent receptor spot, the brightness (number of photons per pixel) and standard deviation of the dot size (σ) are analysed. In addition to the use of STED microscopy, Förster resonance energy transfer (FRET) image spectroscopy and photobleaching step analysis were developed further.
HHU / Cornelia Monzel
In biological cells, the vast majority of functional structures comprise protein molecules. In order to understand the function of proteins, their three-dimensional structure must be known. It is also important to record their spatial arrangement and dynamic interaction amongst themselves and with other proteins in their immediate and wider environment, as these factors frequently trigger their function.
Obtaining this information is very complex and such measurements are in part at the limits of what is technically possible. As the relevant protein structures are often only one to 100 nanometres (nm; one billionth of a metre) in size, they cannot be resolved using conventional optical microscopes. Advanced, high-resolution techniques such as “stimulated emission depletion microscopy” (for short: STED microscopy), which was honoured with the Nobel Prize for Chemistry in 2014, are also not sufficient on their own.
A research team headed by Professor Dr Cornelia Monzel (Institute for Experimental Medical Physics) and Professor Dr Claus A. M. Seidel (Chair of Molecular Physical Chemistry) has now combined various techniques in order to understand the mode of operation of the so-called CD95 receptor on the surface of cell membranes. The aim was to determine the full molecular arrangement and interaction of the receptor required to trigger a cell signal.
The CD95 receptor is of fundamental importance for the cells: The signal, which ultimately leads to controlled or programmed cell death – “apoptosis” – is initiated via this protein. Professor Monzel, one of the two corresponding authors of the study: “On a day-to-day basis, this signalling pathway is responsible for the fact that we do not constantly grow by ensuring that as many existing cells die as new ones are created. It also plays an important role in many other processes such as the development of living beings, wound healing and cancer therapy.”
This receptor is only around 20 nm in size. Dr Nina Bartels, one of the two lead authors of the study, which has now been published in Science Advances: “As we cannot directly and clearly resolve these tiny structures using high-resolution techniques, we had to develop various microscopic and spectroscopic techniques further and combine them.”
In addition to STED microscopy, which can resolve structures down to 40 nm, the researchers in Düsseldorf have also developed the so-called Förster resonance energy transfer (FRET) image spectroscopy and photobleaching step analysis further. As a result, they can measure the number, distribution and interaction of the receptor on the membrane down to just a few nanometres.
Dr Nicolaas van der Voort, the second lead author of the study: “From our measurements, we were able to develop a clear model of which processes are needed to activate the CD95 receptor and thus controlled cell death. To switch on the cell death signal, just 15% of all CD95 receptors on the cell membrane need to bind in twos or threes around a further protein, the so-called CD95 ligand.”
Professor Seidel, the other corresponding author of the study, points to the wider-reaching aspects of the research results: “In addition to the findings relating to the activation of controlled cell death, this work describes important microscopic and spectroscopic advances, which can be transferred to many other biological and medical questions.”
Original publication
Nina Bartels, Nicolaas T. M. van der Voort, Oleg Opanasyuk, Suren Felekyan, Annemarie Greife, Xiaoyue Shang, Arthur Bister, Constanze Wiek, Claus A. M. Seidel, Cornelia Monzel; "Advanced multiparametric image spectroscopy and super-resolution microscopy reveal a minimal model of CD95 signal initiation"; Science Advances, Volume 10