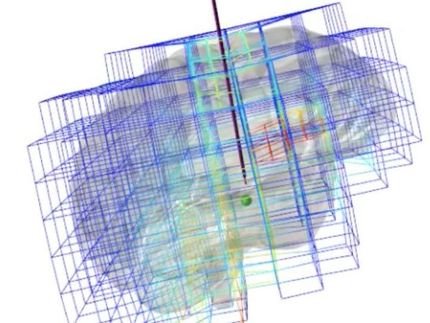
Computer simulation of the plant spindle opens up new possibilities in cell division research
"We were surprised ourselves at how well our simulation reproduced our experimental data"
Advertisement
An interdisciplinary research team led by the Department of Biology at University of Hamburg and the Sainsbury Laboratory in Cambridge (England) has created the first three-dimensional computer simulation of a spindle. This simulation can now be used to better understand fundamental principles of cell division and has been published in the journal "Developmental Cell".
During cell division, the previously duplicated chromosomes, the carriers of the genetic information, must be distributed to the daughter cells in such a way that each daughter cell receives exactly one complete set of chromosomes. The chromosomes are distributed by the spindle, which usually forms in the middle of the dividing cell. Each spindle has two poles, one in each future daughter cell, to which one of the duplicated chromosomes is then moved. Disruptions in this process can lead to severe developmental disorders such as Down's syndrome or cancer.
Dozens of proteins are involved in the formation of the spindle, the function of which are often not or only partially understood. A computer simulation could therefore be used to test whether the data already collected is sufficient to create a complex structure such as the spindle. In addition, other characteristics that are not yet known can be predicted and used as a hypothesis for experimental work.
Many studies on cell division have used the frog Xenopus laevis or the baker's yeast Saccharomyces cerevisiae as model systems. However, the spindle in the frog is very large (40um) and contains approx. 300,000 microtubules. This size makes a computer simulation extremely difficult. In contrast, the spindle in yeast is relatively small (1.4 um) and is formed by only about 40 microtubules. The spindle of Arabidopsis thaliana, a widely used model in molecular plant research, lies between these extremes with a length of approx. 8 um and 2,000 microtubules.
How, an international research team led by Hamburg developmental biologist Prof. Dr. Arp Schnittger from the Department of Biology at University of Hamburg and Dr. François Nedelec from University of Cambridge, in collaboration with other researchers from Germany, England, France and Belgium, has created the first three-dimensional computer simulation of the Arabidopsis spindle and thus of a spindle at all. They were also able to experimentally identify an important regulator of spindle formation.
"We were surprised ourselves at how well our simulation reproduced our experimental data," says Prof. Dr. Arp Schnittger. In the future, Schnittger and his team want to investigate how microtubules attach to chromosomes. "This simulation will be an important tool for us and hopefully also for many colleagues in the plant field and beyond to better understand the regulation of chromosome distribution," concludes Schnittger.
Original publication
Mariana Romeiro Motta, François Nédélec, Helen Saville, Elke Woelken, Claire Jacquerie, Martine Pastuglia, Sara Christina Stolze, Eveline Van De Slijke, Lev Böttger, Katia Belcram, Hirofumi Nakagami, Geert De Jaeger, David Bouchez, Arp Schnittger; "The cell cycle controls spindle architecture in Arabidopsis by activating the augmin pathway"; Developmental Cell