A new method for protection from plant pathogens could help support global food security
By modifying a plant intracellular immune receptor (NLR), researchers have developed a potential new strategy for resistance to rice blast disease, one of the most important diseases threatening global food security. The collaborative team from the UK and Japan have recently published their research in PNAS. This could have implications for future approaches to crop protection and ultimately global food supply stability.
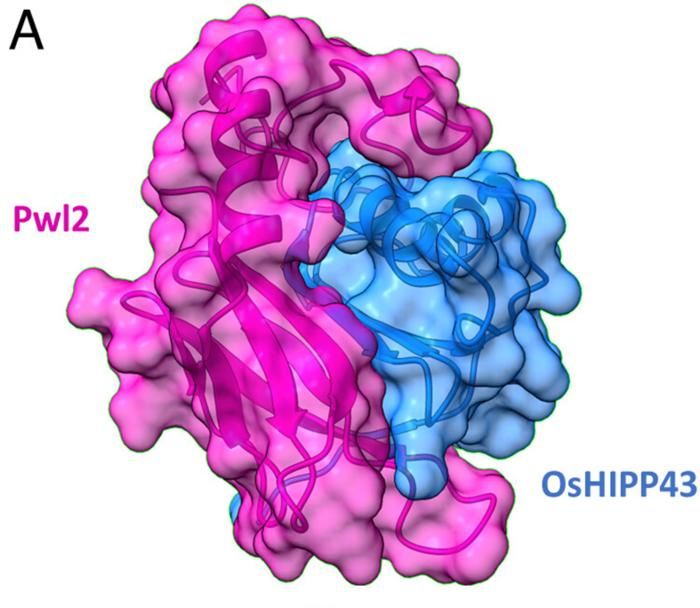
The crystal structure of the complex reveals an extensive interface between Pwl2 and OsHIPP43.
PNAS
The research was led from the Department of Biochemistry and Metabolism at the John Innes Centre, with partners at The Sainsbury Laboratory, University of East Anglia, and the Division of Genomics and Breeding, Iwate Biotechnology Research Center, Japan. For a critical part of the study, the researchers worked with the UK’s national synchrotron, Diamond Light Source.
Rice blast disease remains one of the most recalcitrant diseases threatening global food security. This disease is caused by the filamentous fungus, Magnaporthe oryzae and is directly responsible for the loss of more than 30% of harvested rice annually. This pathogen can also cause blast disease on other cereal crops including wheat and barley.
Current approaches to deployment of durable disease resistance in the field are limited by the pace they can be identified in nature and the evolution of plant pathogens such as the blast fungus that manage to bypass these new resistances. Bioengineering of plant immune receptors such as NLRs has emerged as a new path for generating novel disease resistance traits to counteract the expanding threat of plant pathogens to global food security that can potentially be developed on demand.
Rafał Zdrzałek, the lead author explains “Pathogens secrete proteins called “effectors” into host cells to manipulate plant metabolism and promote infection. Plants can recognise these effectors using immune receptors called NLRs. However, it’s not always easy to define a receptor naturally recognising any given effector, and even if such receptor exists, pathogen’s effectors can mutate and evolve to escape that recognition. Interactions between pathogen effectors and plant receptors are studied to understand the modus operandi of each pathogen, but also allows us to tinker with the natural plant receptors and alter their recognition specificity.”
In their publication the researchers focused on engineering an NLR immune receptor from rice to robustly bind a broader, conserved effector family from the blast fungus pathogen. Mark Banfield, the corresponding author, adds; “By recognising a conserved effector family, this engineered immune receptor establishes a proof-of-principle for future delivery of robust, longer-lived blast disease resistance in agriculture. It may be more difficult for the pathogen to evolve to escape recognition. The concept of host-target immune receptor engineering may also be applicable to other plant diseases that rely on delivery of effectors into host cells for their disease-causing properties.”
By exchanging the heavy metal–associated (HMA) domain of the rice NLR Pikm-1 with that from the rice protein OsHIPP43 (the natural target of the Pwl2 effector), the researchers successfully changed the receptor's response profile to recognise and respond to Pwl2 and the broader Pwl effector family.
The researchers collected X-ray diffraction data at the I04 beamline of the UK’s national synchrotron, Diamond Light Source to study the details of the interaction between these two proteins. The crystal structure of the complex reveals an extensive interface between Pwl2 and OsHIPP43.
The study's findings demonstrate the potential of host target-based NLR engineering in developing new resistance traits that could be less prone to being overcome by pathogen evolution. This research could have far-reaching implications for the future of crop protection and global food supply stability.
Original publication
Rafał Zdrzałek, Yuxuan Xi, Thorsten Langner, Adam R. Bentham, Yohann Petit-Houdenot, Juan Carlos De la Concepcion, Adeline Harant, Motoki Shimizu, Vincent Were, Nicholas J. Talbot, Ryohei Terauchi, Sophien Kamoun, Mark J. Banfield; "Bioengineering a plant NLR immune receptor with a robust binding interface toward a conserved fungal pathogen effector"; Proceedings of the National Academy of Sciences, Volume 121, 2024-7-5





















































