Blueprint for blueberry improvement: genetic and epigenetic discoveries
Recent research has uncovered significant genetic and epigenetic variations in blueberry cultivars, particularly between northern highbush (NHB) and southern highbush (SHB) blueberries. The study highlights gene introgression's role in SHB's adaptation to subtropical climates and identifies key genes, such as VcTBL44, associated with fruit firmness. These findings offer valuable insights and resources for future blueberry breeding.
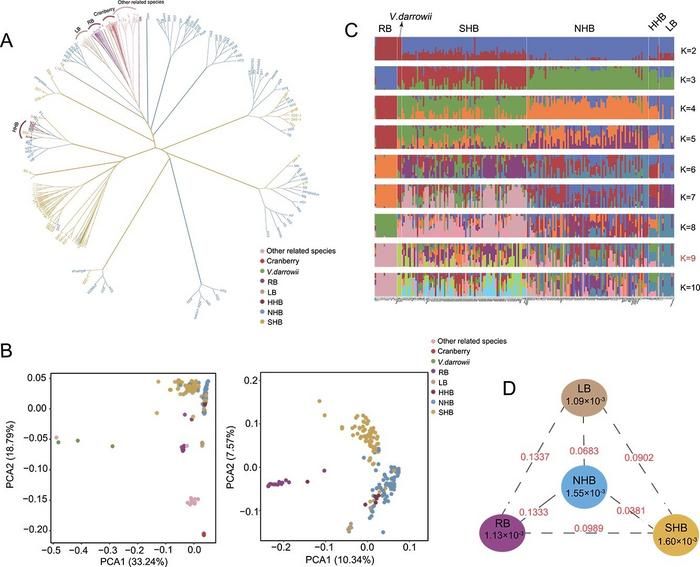
Population structure and genetic diversity of blueberry accessions. A Maximum likelihood phylogenetic tree of 222 blueberry accessions. HHB: half-high blueberry; LB: lowbush blueberry; NHB: northern highbush blueberry; RB: rabbiteye blueberry; SHB: southern highbush blueberry. B Principal component analysis of the first two components (PC1 and PC2) for all accessions and major cultivated blueberry accessions. PC1, first principal component; PC2, second principal component. C Population structure analysis of cultivated blueberry accessions and V. darrowii given different cluster numbers (K = 2–10). The y axis quantifies subgroup membership, and the x axis shows the different accessions. The K value marked in red is the optimal K value determined based on the CV error. D Nucleotide diversity (π) and population divergence (FST value) across four cultivated blueberry subgroups (calculated from the diploid model). The values in the circles represent the nucleotide diversity (π) of the groups (brown, orange, blue, and purple circles represent the LB, SHB, NHB, and RB subgroups, respectively), and the value between each pair indicates population divergence (FST value).
Horticulture Research
Blueberries, part of the Vaccinium genus, are renowned for their nutritional benefits and increasing global demand. However, cultivation faces challenges like climate adaptability and fruit quality. Modern blueberries have a short domestication history, primarily through interspecific hybridization. These challenges necessitate deeper research into the genetic and epigenetic factors influencing blueberry traits.
Researchers from Peking University and Jilin Agricultural University, in collaboration with international experts, have made significant strides in blueberry genetic research. Their study presents a comprehensive analysis of blueberry genomic variation, marking a pivotal moment in agricultural science.
The study involved whole-genome re-sequencing and bisulfite sequencing on various blueberry cultivars to understand their genetic and epigenetic differences. Researchers identified significant gene introgression from V. darrowii and V. ashei into southern highbush (SHB), aiding its subtropical adaptation. They discovered the VcTBL44 gene, crucial for regulating fruit firmness in SHB. Additionally, they found significant differences in DNA methylation patterns between northern highbush (NHB) and SHB, particularly in CHH-DMRs associated with transposon regulation. These findings offer a comprehensive understanding of the genetic and epigenetic mechanisms that have improved blueberry cultivars, providing valuable resources for future breeding programs aimed at enhancing fruit quality and climate resilience.
Dr. Haiyue Sun, a leading researcher in the study, stated, "Our research provides a detailed genetic and epigenetic map of blueberries, offering crucial insights for breeding programs. The identification of key genes like VcTBL44 paves the way for developing cultivars with improved fruit quality and climate adaptability."
The insights from this study have significant implications for blueberry breeding. The genetic and epigenetic resources identified can develop new cultivars more resilient to climate changes and superior in fruit quality. This research enhances our understanding of blueberry genetics and provides practical tools for breeders to meet the growing consumer demand for high-quality blueberries.
Original publication
Zejia Wang, Wanchen Zhang, Yangyan Zhou, Qiyan Zhang, Krishnanand P Kulkarni, Kalpalatha Melmaiee, Youwen Tian, Mei Dong, Zhaoxu Gao, Yanning Su, Hong Yu, Guohui Xu, Yadong Li, Hang He, Qikun Liu, Haiyue Sun; "Genetic and epigenetic signatures for improved breeding of cultivated blueberry"; Horticulture Research, Volume 11, 2024-5-14