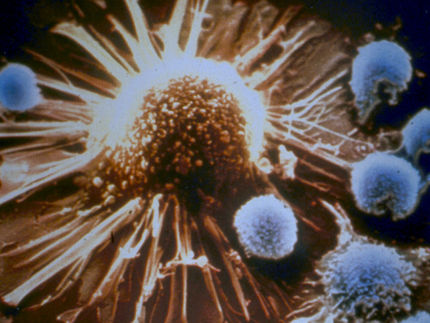
How immune cells recognize the abnormal metabolism of cancer cells
New targets for cancer immunotherapies?
When cells become tumor cells, their metabolism changes fundamentally. Researchers at the University of Basel and the University Hospital Basel have now demonstrated that this change leaves traces that could provide targets for cancer immunotherapies.
Cancer cells function in turbo mode: Their metabolism is programmed for rapid proliferation, whereby their genetic material is also constantly copied and translated into proteins. As researchers led by Professor Gennaro De Libero from the University of Basel and the University Hospital Basel now report, this turbo metabolism leaves traces on the surface of tumor cells that can be read by specific immune cells. The research team's findings have been published in the journal Science Immunology.
The immunologists working with De Libero discovered the immune cells in question, known as MR1T cells, around ten years ago. This previously unknown type of T cell can attack and eliminate tumor cells. Since then, the team has been researching these cells as a potential tool for novel immunotherapies against a variety of different types of cancer.
Modified DNA and RNA building blocks
The team has now been able to decipher exactly how the T cells recognize the degenerated cells: The altered metabolism of the cancer cells produces a certain type of molecule that appears on the surface of these degenerated cells. “These molecules are chemically modified DNA and RNA building blocks that are the result of changes in three important metabolic pathways,” explains De Libero.
“The fact that cancer cells have a profoundly altered metabolism makes them recognizable to MR1T cells,” adds Dr. Lucia Mori, who was involved in the research. In previous work, the researchers had already discovered that these T cells recognize a surface protein found on all cells, known as MR1. It acts as the proverbial silver platter and presents metabolic products from inside the cell on the cell surface so that the immune system can then check to see whether the cell is healthy or not.
“Several metabolic pathways are altered in cancer cells. This produces particularly suspicious metabolic products and thus alert the MR1T cells,” explains Dr. Alessandro Vacchini, the study's first author.
As a next step, the researchers intend to examine in more detail just how these telltale metabolites interact with the MR1T cells. The long-term vision: Within the framework of future therapies, a patient’s T cells could be reprogrammed and optimized to recognize and attack these cancer-typical molecules.
Original publication
Alessandro Vacchini, Andrew Chancellor, Qinmei Yang, Rodrigo Colombo, Julian Spagnuolo, Giuliano Berloffa, Daniel Joss, Ove Øyås, Chiara Lecchi, Giulia De Simone, Aisha Beshirova, Vladimir Nosi, José Pedro Loureiro, Aurelia Morabito, Corinne De Gregorio et al.; "Nucleobase adducts bind MR1 and stimulate MR1-restricted T cells"; Science Immunology, Volume 9
Most read news
Original publication
Alessandro Vacchini, Andrew Chancellor, Qinmei Yang, Rodrigo Colombo, Julian Spagnuolo, Giuliano Berloffa, Daniel Joss, Ove Øyås, Chiara Lecchi, Giulia De Simone, Aisha Beshirova, Vladimir Nosi, José Pedro Loureiro, Aurelia Morabito, Corinne De Gregorio et al.; "Nucleobase adducts bind MR1 and stimulate MR1-restricted T cells"; Science Immunology, Volume 9
Topics
Organizations
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.