Mix and Match: How a Methanogen creates its own sulfate reduction machinery
Discovery opens up exciting opportunities in biofuel production
Advertisement
Scientists at the Max Planck Institute for Marine Microbiology in Bremen, Germany, have uncovered the molecular secrets of a methane-generating microbe that can transform sulfate into sulfide – a ready-to-use cellular building block. This discovery opens up exciting opportunities in biofuel production.
PhD-student Marion Jespersen works on a fermenter in which M. thermolithotrophicus grows exclusively on sulfate as sulfur source.
Tristan Wagner, Max-Planck-Institut für Marine Mikrobiologie
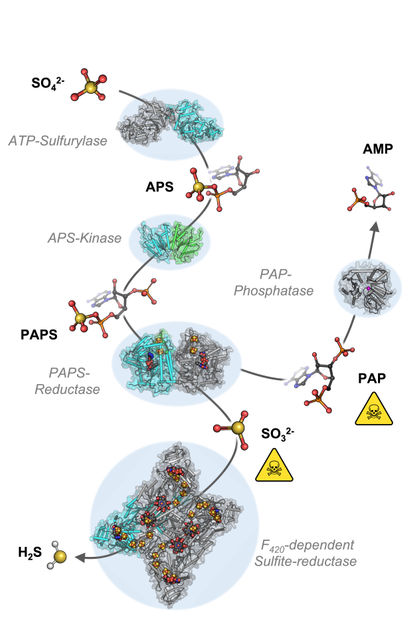
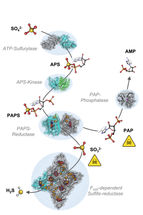
The cascade of chemical reaction starting from sulfate (SO42-) to sulfide (H2S).
Marion Jespersen, Max-Planck-Institut für Marine Mikrobiologie

Sulfur, an essential building block of life
Sulfur is a fundamental element of life and all organisms need it to synthesize cellular materials. Autotrophs, such as plants and algae, acquire sulfur by converting sulfate into sulfide, which can be incorporated into biomass. However, this process requires a lot of energy and produces harmful intermediates and byproducts that need to be immediately transformed. As a result, it was previously believed that microbes known as methanogens, which are usually short on energy, would be unable to convert sulfate into sulfide. Therefore, it was assumed that these microbes, which produce half of the world's methane, rely on other forms of sulfur, such as sulfide.
A methanogen assimilating sulfate?
This dogma was broken in 1986 with the discovery of the methanogen Methanothermococcus thermolithotrophicus, growing on sulfate as the only sulfur source. How is this possible, considering the energetic costs and toxic intermediates? Why is it the only methanogen that seems to be capable of growing on this sulfur species? Does this organism use chemical tricks or a yet unknown strategy to allow sulfate assimilation? Marion Jespersen and Tristan Wagner at the Max Planck Institute for Marine Microbiology have now found answers to these questions and published them in the journal Nature Microbiology.
The first challenge the researchers met was to get the microbe to grow on the new sulfur source. “When I started my PhD, I really had to convince M. thermolithotrophicus to eat sulfate instead of sulfide,” says Marion Jespersen. “But after optimizing the medium, Methanothermococcus became a pro at growing on sulfate, with cell densities comparable to those when growing on sulfide.”
“Things got really exciting when we measured the disappearance of sulfate as the organism grew. This was when we could really prove that the methanogen converts this substrate.” This allowed the researchers to safely cultivate M. thermolithotrophicus in bioreactors in large scales, as they were no longer dependent on the toxic and explosive hydrogen sulfide gas for growth. “It provided us with enough biomass to study this fascinating organism,” explains Jespersen. Now the researchers were ready to dig into the details of the underlying processes.
The first molecular dissection of the sulfate assimilation pathway
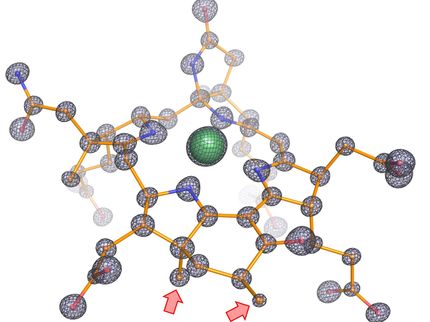
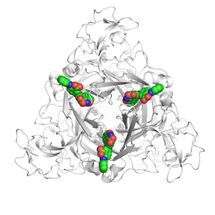
To understand the molecular mechanisms of sulfate assimilation, the scientists analyzed the genome of M. thermolithotrophicus. They found five genes that had the potential to encode sulfate-reduction-associated enzymes. “We managed to characterize every one of those enzymes and therefore explored the complete pathway. A true tour de force when you think about its complexity,” says Tristan Wagner, head of the Max Planck Research Group Microbial Metabolism.
By characterizing the enzymes one-by-one, the scientists assembled the first sulfate assimilation pathway from a methanogen. While the first two enzymes of the pathway are well known and occur in many microbes and plants, the next enzymes were of a new kind. “We were stunned to see that it appears as if M. thermolithotrophicus has hijacked one enzyme from a dissimilatory sulfate-reducing organism and slightly modified it to serve its own needs,” says Jespersen. While some microbes assimilate sulfate as a cellular building block, others use it to obtain energy in a dissimilatory process – as humans do when respiring oxygen. The microbes that perform dissimilatory sulfate-reduction employ a different set of enzymes to do so. The methanogen studied here converted one of these dissimilatory enzymes into an assimilatory one. “A simple, yet highly effective strategy and most likely the reason why this methanogen is able to grow on sulfate. So far, this particular enzyme has only been found in M. thermolithotrophicus and no other methanogens,” Jespersen explains.
However, M. thermolithotrophicus also needs to cope with two poisons that are generated during the assimilation of sulfate. That´s what the last two enzymes of the pathway are made for: The first one, again similar to a dissimilatory enzyme, generates sulfide from sulfite. The second one is a new type of phosphatase with robust efficiency to hydrolyze the other poison, shortly known as PAP.
“It seems that M. thermolithotrophicus collected genetic information from its microbial environment that enabled it to grow on sulfate. By mixing and matching assimilatory and dissimilatory enzymes, it created its own functional sulfate reduction machinery,” says Wagner.
New avenues for biotechnological application
Hydrogenotrophic methanogens, such as M. thermolithotrophicus, have the amazing ability to convert dihydrogen (H2, for example artificially produced from renewable energy) and carbon dioxide (CO2) into methane (CH4). In other words, they can convert the greenhouse gas CO2 into the biofuel CH4, which can be used, for example, to heat our homes. To do this, methanogens are grown in large bioreactors. A current bottleneck in the cultivation of methanogens is their need for the highly hazardous and explosive hydrogen sulfide gas as a sulfur source. With the discovery of the sulfate-assimilation pathway in M. thermolithotrophicus, it is possible to genetically engineer methanogens that are already used in biotechnology to use this pathway instead – leading to safer and more cost-effective biogas production.
“An unresolved burning question is why M. thermolithotrophicus would assimilate sulfate in nature. For this, we will have to go out into the field and see if the enzymes required for this pathway are also expressed in the natural environment of the microbe”, concludes Wagner.