Primate DNA reveals applications to human health
Genomes of 233 primate species reveal key features of human evolution, human disease and biodiversity conservation
New research led by Tomàs Marquès-Bonet, ICREA Researcher at IBE (CSIC-UPF) and Professor of Genetics in the Department of Medicine and Life Sciences (MELIS) at Pompeu Fabra University (UPF); Dr. Kyle Farh(Illumina) and Dr. Jeffrey Rogers (Baylor College of Medicine), combines genome sequencing of more than 809 individuals from 233 primate species, covering almost half of all existing primate species on Earth, covering almost half of all existing primate species on Earth. Jeffrey Rogers (Baylor College of Medicine), combines the genome sequencing of more than 809 individuals from 233 primate species, covering almost half of all primate species existing on Earth, including the study of fossil remains and multiplying by 4 the number of primate genomes available so far.
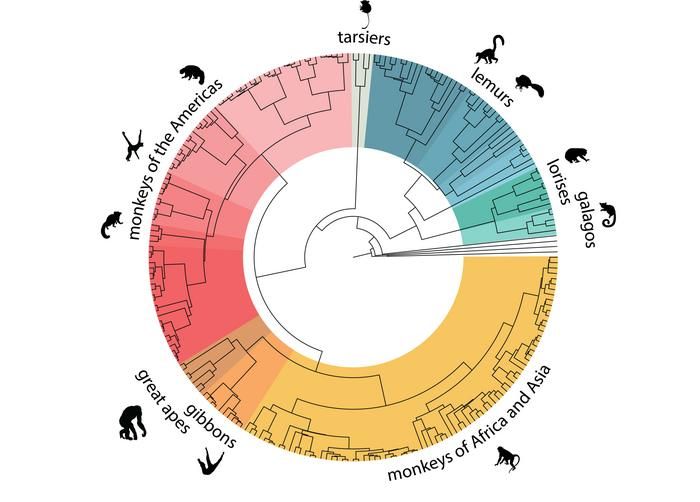
Primates phylogenetic tree.
Lukas Kuderna
The study provides new information on the genetic diversity and phylogeny of primates, which is important for understanding and conserving the diversity of the species closest to our own. Similarly, it identifies a new way to find genetic mutations that could be responsible for many human diseases.
Rare mutations may increase disease risk
One of the limitations of human and clinical genetics is the current inability to detect, among hundreds of thousands of mutations, those that cause disease. The genetic causes of many common diseases, such as diabetes and heart disease, are currently unknown due to the lack of genetic information or the large number of genetic factors involved. Some diseases are thought to originate when a set of genetic variations or mutations with a "mild" effect act together to cause a disease of polygenic origin, such as diabetes or cancer.
By comparing the genomes of 809 individuals from 233 non-human primate species with the human genome, the research has identified 4.3 millionmissensemutations that affect amino acid composition and can alter protein function. These mutations identified in primates are the basis for comparative studies with human variants and to identify key variants in many human diseases.
"Six percent of these mutations are abundant in primates and are therefore considered "potentially benign" in human diseases, since their presence is tolerated in these animals," says Kyle Farh, vice president of Artificial Intelligence at Illumina and co-lead author of the publications.
The identification of disease-causing mutations has been achieved thanks to the PrimateAI-3D deep learning algorithm. An artificial intelligence (AI) algorithm developed by Illumina, the world's leading DNA sequencing company, which is a kind of ChatGPT for genetics that uses genome sequence instead of human language.
New insights into primate evolution and human uniqueness
The publication of this project contains the most comprehensive catalog of primate genomic information to date, covering nearly half of all primate species on Earth. It contains information on primates from Asia, America, Africa and Madagascar.
According to Tomàs Marquès-Bonet, "humans are primates. The study of hundreds of non-human primate genomes, given their phylogenetic position, is very valuable for human evolutionary studies, to better understand the human genome and the basis of our uniqueness, including the basis of human diseases, and for their future conservation".
These studies have also indicated that primate genetics do not always correspond to their taxonomy. We found several cases in which relationships between primate species are described more as complex networks than as simple branching trees.
Another study delves into the evolution of baboon species, a large and diverse group of monkeys, showing that there have been several episodes of hybridization and gene flow between species that had not previously been recognized. In addition, we found that the yellow baboons of western Tanzania are the first non-human primates to have received genetic input from three distinct lineages. "These results suggest that the population genetic structure and history of introgression between baboon lineages is more complex than previously thought, and that demonstrates that baboons are a good model for the evolution of humans, Neanderthals and Denisovans." Says Jeffrey Rogers, associate professor in the Human Genome Sequencing Center and the Department of Molecular and Human Genetics at Baylor College of Medicine, who co-led this study.
"Our studies provide clues as to which species are most urgently in need of conservation efforts, and may help identify the most effective strategies to preserve them," says Lukas Kuderna, first author of one of the studies.
Finally, the new genomic catalog has halved the number of genomic innovations that were thought to be exclusively human. This observation facilitates the identification of those mutations not shared with primates that may therefore be unique to human evolution and the characteristics that make us human.
Note: This article has been translated using a computer system without human intervention. LUMITOS offers these automatic translations to present a wider range of current news. Since this article has been translated with automatic translation, it is possible that it contains errors in vocabulary, syntax or grammar. The original article in Spanish can be found here.