Systematic depiction of previously unknown gene functions
Largest cellular proteomic study of yeast cells unlocks fundamental insights
A proteome is the entirety of all active protein molecules present within an organism, a certain type of tissue, or a cell under specific conditions and at a certain time. Now, an international team of researchers led by members from Charité – Universitätsmedizin Berlin, the Francis Crick Institute in London, the University of Edinburgh and the University of Zürich has constructed the largest cellular proteomic map to date, using yeasts as model organisms. It offers insight into previously unstudied genes and the ways in which proteins are expressed and regulated based on their encoding. The study appears in the latest issue of the journal Cell.
Despite decades of research, the function of many genes remains unknown, limiting our understanding of certain diseases, some of them rare, and hindering the development of new therapies. In the case of yeast and bacterial cells, there is a lack of the fundamental knowledge required to develop the new antimicrobials that are urgently needed in the face of rising drug tolerance and resistance. The current study is one of the most extensive proteomic studies in the world. The team of researchers studied yeast cells to gain a more accurate and detailed picture of the role of genes that were previously not yet associated with specific functions. The goal is to obtain critical information to interpret the ramifications of genetic mutations and help to close diagnostic gaps. The researchers’ objective was to investigate in detail how certain proteins are expressed and regulated, which should ultimately also help lay the groundwork for developing new drugs.
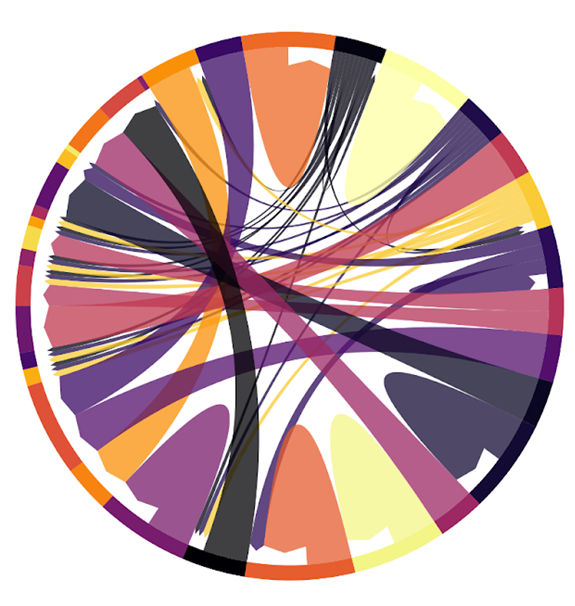
Visual summary of how the removal of all non-essential genes affects the behavior of the remaining budding yeast proteome. Each arrow on the plot represents a different biological process (a 'Gene Ontology'), and connects the deleted gene to responding genes. This information can help researchers understand how different genes and processes interact with each other, and can provide insights into the process a thus far unknown gene is involved in.
© Charité | Institut für Biochemie
Dr. Markus Ralser, senior author of the study and Director of the Institute of Biochemistry and Einstein Professor of Biochemistry at Charité, explains: “We made use of a collection of yeast strains generated by an international consortium, in which all nonessential genes were missing from at least one strain. We then used mass spectrometry, a technology that can quantify thousands of proteins in parallel, to characterize each of the strains, resulting in the largest proteomic study to date.” The study revealed general principles governing the expression of proteins, including identifying for thousands of proteins how their function or biophysical properties relate to expression.
The research also generated a lot of data on previously understudied proteins. This resulted in the development of new methods for assigning gene function. Mass spectrometry, and especially specific proteomic techniques developed by Ralser and his team in recent years, helped to determine the amounts of specific proteins present in the individual yeast strains in the absence of all nonessential genes. “The research has the potential to fundamentally improve our understanding of cell biology, providing new insights into gene function in life forms whose cells have a nucleus, which are known as eukaryotes,” says Dr. Georg Kustatscher, who contributed to the study by analyzing the large dataset with his research group at the University of Edinburgh. “The proteomes recorded in the study contain key information about potential new drug targets, providing hope for future treatment options.”
The extensive study was made possible by a collaboration between research teams at Charité, the Francis Crick Institute, and the University of Edinburgh. Labs at Cambridge and the University of Toronto also contributed by pioneering essential proteomic technologies and functional genomics methods used in the study. Dr. Christoph Messner, who was initially responsible for the study at the Francis Crick Institute in London and is now a group leader at the University of Zurich, points out: “To our surprise, the study further revealed that the response of a protein to these mutations depends more on its biophysical properties than its function. This opens up a new way of looking at analysis of large biological datasets, which these days are often already collected using advanced sequencing or mass spectrometry techniques, but can still be difficult to interpret.” The researchers thus conclude that their study will have far-reaching implications for the biosciences.
This research has provided critical information on gene function and protein expression, paving the way for future breakthroughs in the field of microbiology. The team is now working on conducting a similar study in human cells, with the goal of generating more information on previously unknown genes. The researchers also aim to link the yeast proteome maps with other molecular data to help develop better antifungal therapies.
Original publication
Other news from the department science
Most read news
More news from our other portals
See the theme worlds for related content
Topic World Mass Spectrometry
Mass spectrometry enables us to detect and identify molecules and reveal their structure. Whether in chemistry, biochemistry or forensics - mass spectrometry opens up unexpected insights into the composition of our world. Immerse yourself in the fascinating world of mass spectrometry!
Topic World Mass Spectrometry
Mass spectrometry enables us to detect and identify molecules and reveal their structure. Whether in chemistry, biochemistry or forensics - mass spectrometry opens up unexpected insights into the composition of our world. Immerse yourself in the fascinating world of mass spectrometry!