Scientific image analysis for everyone
New software enables automated analysis of image data without programming knowledge
The software JIPipe was developed by scientists at the Leibniz Institute for Natural Product Research and Infection Biology (Leibniz-HKI) and significantly simplifies the analysis of images generated in research. Users can create flowcharts according to their application needs and thus perform automatic image analyses using artificial intelligence without any programming knowledge. JIPipe is based on ImageJ, a standard program for scientific analysis of biomedical microscopic images. The authors now present their development in Nature Methods.
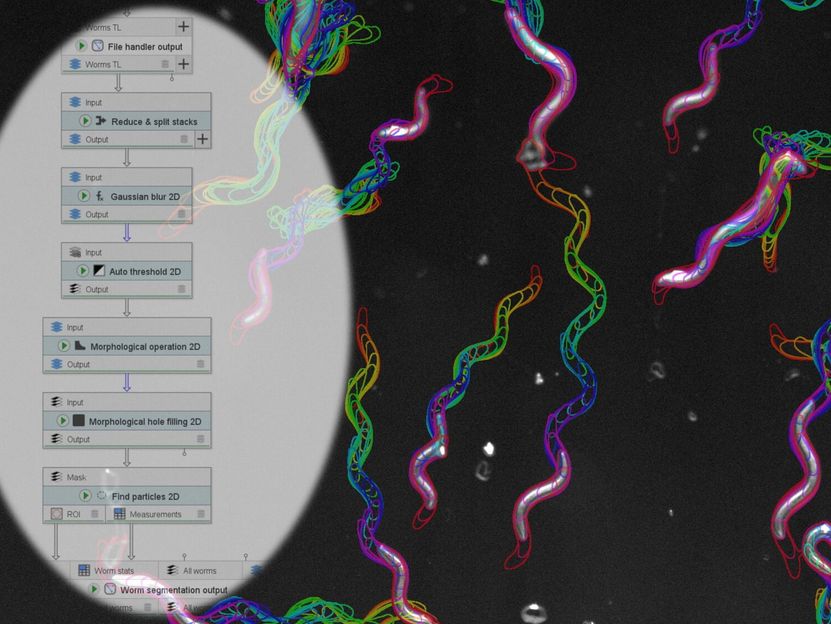
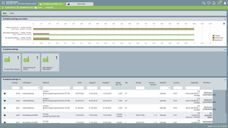
JIPipe can be used, for example, to analyse the movement profiles of nematodes.
Hannah Büttner, Zoltán Cseresnyés, Ruman Gerst
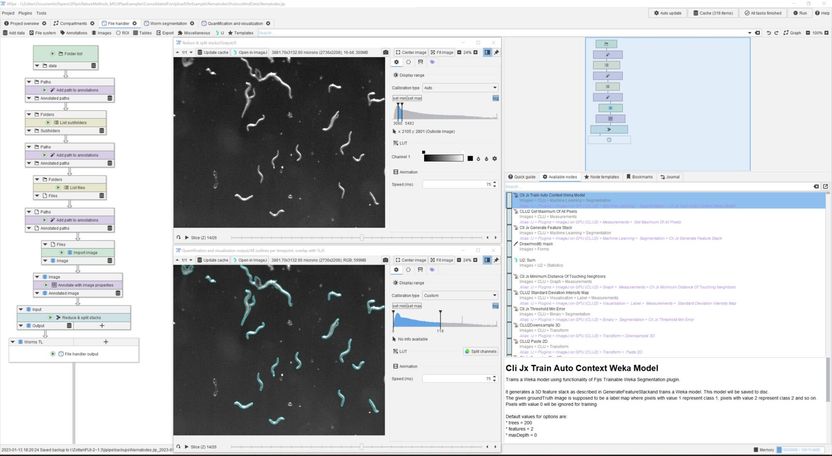
Screenshot of a JIPipe analysis of nematode survival.
Zoltán Cseresnyés/Leibniz-HKI
Images, especially microscopic images, play a major role in biomedical research. With the help of fluorescent labels, for example, processes in cells become visible. "A picture is worth a thousand words - that is still true," says Thilo Figge, head of the Applied Systems Biology research group at Leibniz-HKI and professor at Friedrich Schiller University Jena. But analysis poses increasing challenges for researchers. "Higher and higher resolutions and thus larger amounts of data are being generated," Figge explains. "At the same time, the methods of AI, or artificial intelligence, are now so advanced that they are increasingly difficult to use for researchers without programming skills."
The open source program JIPipe - short for Java Image Processing Pipeline - developed at Leibniz-HKI aims to simplify that. "JIPipe is a tool that does not require any programming skills," explains developer Ruman Gerst, a member of the Applied Systems Biology research group. Instead, the software uses a visual programming language: with the help of prefabricated building blocks, users can create individual workflows to automatically analyze images according to their specific requirements.
JIPipe supports other programming languages
The program is based on the open source software ImageJ, which has established itself as a standard in scientific image analysis. JIPipe and ImageJ are fully compatible and complement each other in scientific image analysis. "Our program supports ImageJ scripts and includes the usual functions and macros," Gerst explains. Other programming languages such as Python and R are also supported.
The precursor program was developed several years ago by Zoltán Cseresnyés, also a member of the Applied Systems Biology research group. "Originally, I wrote the code for a phagocytosis assay," Cseresnyés explains. In phagocytosis, a cell takes up a particle, such as another cell, and breaks it down, which is usually visualized with fluorescent dyes.
Over time, the imaging specialist kept extending the code for new applications, and the program became unwieldy and too complex. "We realized we had to redesign it and make it modular," Cseresnyés says, which is why the team brought bioinformatician Ruman Gerst on board. He also suggested the current visual programming language, which allows the team to analyze image data from any biomedical problem.
Reproducible results
JIPipe has already been used for several studies, for example to investigate the efficiency of drug delivery by so-called nanocarriers in the liver or to test the survival rate of nematodes that have digested toxin-producing bacteria. Confrontations between immune cells and fungal spores have also been analyzed with the new program. The developers also offer courses for use as part of the Microverse Imaging Center and the NFDI4BioImage, part of the German National Research Data Infrastructure. "In contrast to manual image analysis, automated analysis always provides the same results, is therefore reproducible and complies with the so-called FAIR principles for image analysis," Thilo Figge emphasizes. The term FAIR originates in the field of research data management and stands for "Findable, Accessible, Interoperable and Reusable".