Zombie viruses on a hijacking trip
Ancient, dormant sequences in the genome impact embryonic development in unexpected ways
The mammalian genome contains retroviral sequences that are in an undead but mostly “harmless” state. An international research team recently discovered how some of these retroviral gene fragments affect embryonic cells if they are unleashed. Unexpectedly, not the viral proteins, but rather copies of the genetic material itself generate an imbalance in the cell.

Symbolic image
Computer-generated image
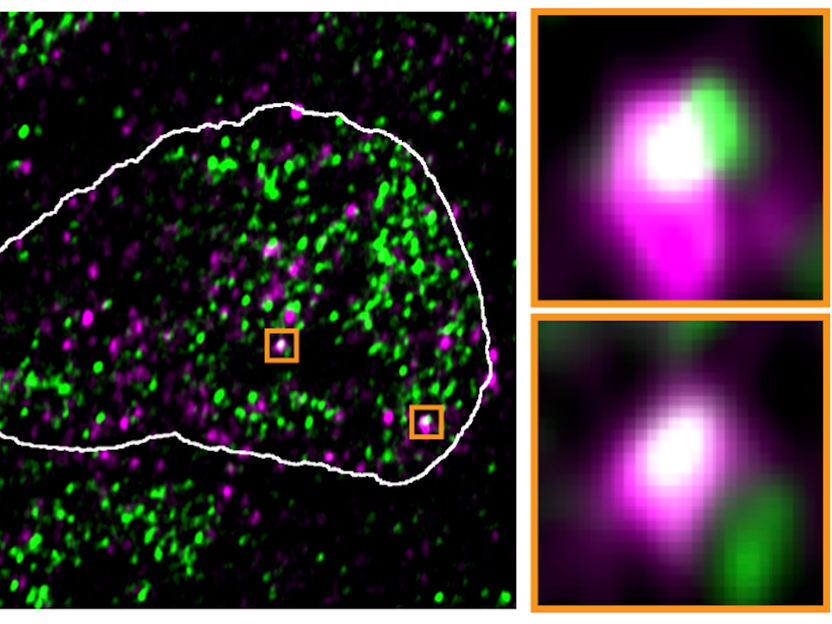
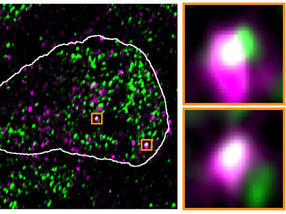
In the nucleus of mouse stem cells, derepressed endoviral RNA molecules (purple) appear at the same spots as condensates containing RNA polymerase II (green), as this fluorescence microscopic image shows
Asimi et al., MPIMG

Over thousands of years of evolution, countless viruses have embedded themselves in our genome. A staggering ten percent of mammalian genomes consist of ancient retroviral sequences. These no longer seem to pose any danger, because most of them have mutated beyond recognition. Additionally, these genes have been epigenetically silenced by the cell. But as the silencing of the viral remains fails, they will rise from their graves, causing chaos in the cell.
“We found that the messenger copies of some of the viral genes, the RNA, have an important impact on embryonic cells,” says Denes Hnisz, research group leader at the Max Planck Institute for Molecular Genetics (MPIMG) in Berlin. “The viral sequences seem to remember their original mission of hijacking the molecular machinery that ensures the flow of information from DNA to RNA to protein. Interestingly, the messenger RNA itself seems to be responsible.”
Hnisz’ team and collaborating researchers published their results in the journal Nature Genetics. They describe that the RNA of the resurrected viruses exerts attractive forces on the enzymes that read the information from the DNA. The tasks of the embryonic cell – such as reading important embryonic genes – are neglected and a fatal imbalance develops. This unleashed state occurs, for example, in some types of cancer and neurological diseases.
Mostly harmless viral remains
Viruses are cleverly constructed snippets of genetic information. Some of them incorporate themselves into the genome of their hosts and persist there. Thousands of copies of Endogenous Retroviruses (ERVs) have spread throughout mammalian genomes, often in droves of hundreds of repetitive copies.
“As retroviruses jump from one section of DNA to the next during their life cycle, they can alter genes and even recombine them. This makes them an important tool for evolution to create new genes,” says Henri Niskanen, one of the scientists involved in the study. “For an individual organism however, uncontrolled gene modification does not bode well, especially during embryonic development.”
This is why the cell will identify ERV sequences and recruit dedicated repressive machinery to their sites and keep them silent. Additionally, the chromosome is getting compacted at these sites.
Resurrected viruses hijack transcriptional machinery
But what happens if you turn off these protective mechanisms? The research team wanted to find out what is the very first thing that happens when the ERV zombies are no longer kept in check. For this purpose, they removed Trim28, a protein that is responsible for silencing the viral remnants, from embryonic stem cells of mice, and monitored the immediate consequences.
Once Trim28 was gone, the cell unsurprisingly read more ERV genes, producing RNA copies with the help of the RNA polymerase enzyme. But unexpectedly, the polymerase simultaneously disappeared from stem cell genes that are especially important for stem cell potency.
“Only a limited pool of polymerase enzymes and other required factors is available in each cell,” says Christina Riemenschneider, another researcher of the team. If too many genes are transcribed at the same time, they will compete for the limited resources, she says. In an experiment, repeats of ERV sequences competed against stem cell genes. “We see that ERV repeats have a slightly higher affinity – they draw the machinery away from embryonic genes, creating an imbalance,” says Riemenschneider.
Attractive forces between molecules
RNA polymerase and other necessary factors that selectively dock onto genes often assemble into droplets that contain a lot of proteins and float around in the cell nucleus – much like oil droplets in a salad dressing. These “condensates” contain many of the molecules necessary for reading genes and they are particularly attracted to specific DNA segments that control the most important genes in a cell.
The ERV genes, or rather the RNA molecules produced using these genes, seemed to literally hijack the condensates. In high-resolution microscopic images, they were often in the same locations as the reactivated ERV genes. After the viral RNA was removed from the cells, the droplets returned to their original location.
The effects of the virus-like RNA were not limited to the molecular level. Working in early mouse embryos, the research team demonstrated that the shift of the condensates toward the ERVs had adverse effects on development. Stem cells, for example, lost their typical property of being able to develop into any other cell because necessary genes were no longer active.
Relevant to evolution and the development of disease
“It's quite remarkable that non-coding, non-functional genes have such a profound effect via RNA,” says scientist Abhishek Sampath Kumar, who was involved in the work. “You might imagine DNA damage or viral particles when thinking of retroviruses that integrate into the genome, but that is not the case in this instance.”
As a result, the team of scientists says their finding puts research on endogenous retroviruses in a new light. “Hijacking of transcriptional condensates by the ERVs and their RNA is an important mechanistic finding that should be taken into account in future studies of transposable elements and their epigenetic regulators,” says researcher Vahid Asimi, who worked on the study. “This could be an additional route ERVs used to contribute to evolutionary innovation.”
“Reactivation of ERVs is clearly linked to pathologies, from obesity to various cancers to neurological diseases such as amyotrophic lateral sclerosis and schizophrenia,” adds group leader Denes Hnisz. “Hopefully, our research will help elucidate the molecular causes of these diseases.”
Original publication
Asimi V, Sampath Kumar A, Niskanen H, Riemenschneider C, Hetzel S, Naderi J, Fasching N, Popitsch N, Du M, Kretzmer H, Smith ZD, Weigert R, Walther M, Mamde S, Meierhofer D, Wittler L, Buschow R, Timmermann B, Cisse II, Ameres SL, Meissner A, Hnisz D. Hijacking of transcriptional condensates by endogenous retroviruses. Nat Genet. 2022 Aug;54(8):1238-1247.