Bird’s enzyme points toward novel therapies
Rice University scientists imbue cells with ‘noncanonical’ pathway to make own drugs
Thank the rare crested ibis for a clue that could someday help our bodies make better drugs.

Thank the rare crested ibis for a clue that could someday help our bodies make better drugs (symbolic image).
Computer generated picture
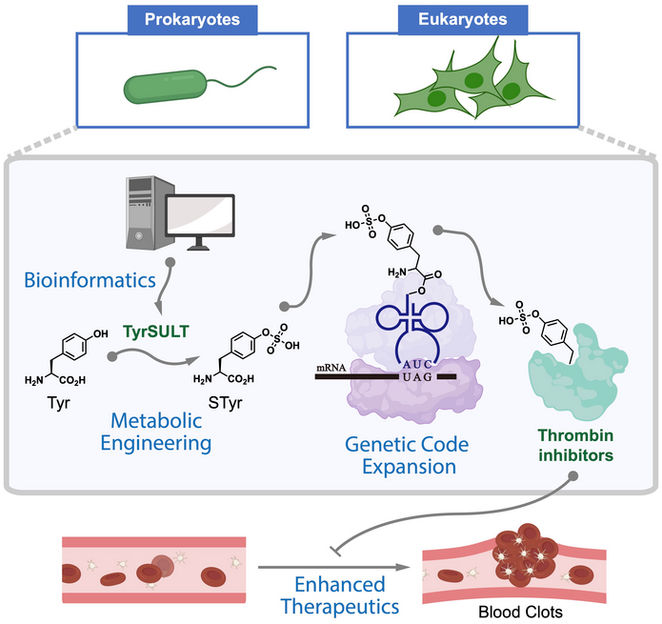
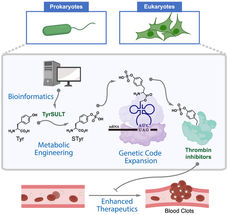
A graphic shows how Rice University chemists used a rare genetic pathway to metabolically engineer cells that serve as drug factories to make thrombin inhibitors that break up blood clots. The study began with a bioinformatic survey that found the key in a crested ibis.
Xiao Lab/Rice University
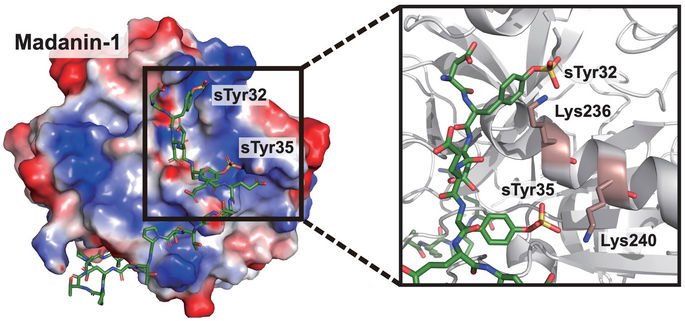
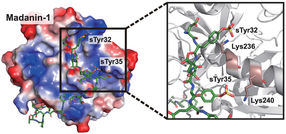
Rice University scientists developed cells engineered to express therapeutic proteins, specifically a thrombin inhibitor. The key is the site-specific insertion of sulfotyrosine (sTyr), a mutant of the standard amino acid tyrosine found naturally only in the crested ibis.
Xiao Lab/Rice University

The species of bird is the only one known to naturally produce an enzyme able to generate a noncanonical amino acid; that is, one not among the 20 necessary to encode most proteins.
That it exists -- a discovery made through computational comparison of genome databases -- proves it’s possible for that enzyme to work within the context of living cells, even if scientists don’t know what it does for the bird.
But they have a pretty good idea of what it could do for us.
A new study by Rice University chemist Han Xiao, theoretical physicist Peter Wolynes and their colleagues shows that amino acid, sulfotyrosine (sTyr), a mutant of the standard amino acid tyrosine, is a key building block to program living cells that express therapeutic proteins. It could potentially allow cells to serve as sensors that monitor their environments and respond with the necessary treatment.
Mimicking the ibis’ ability to synthesize sTyr and incorporate it into proteins requires modifying a cell’s DNA with a mutant codon that, in turn, makes the transferase enzyme, sulfotransferase 1C1, found in the bird. This catalyzes the generation of sTyr, an essential recognition moiety in a variety of biomolecular interactions.
The proof-of-concept study produced for the first time mammalian cells that synthesize sTyr. In an experiment, the Xiao lab made cells that enhanced the potency of thrombin inhibitors, anticoagulants used to prevent blood clotting.
The study appears in Nature Communications.
“In nature, most of our species are made with 20 canonical building blocks,” Xiao said. “If you want to add an additional building block, you need to think about how to make it. We solved that problem: We can ask the cell to make it.
“But then we have to have the translational machinery to recognize it. And a special codon to encode this new building block,” he said. “With this study, we’ve fulfilled all three of these requirements.”
Xiao received a National Institutes of Health grant in 2019 to see if cells could be programmed to make substances with extra amino acids. The new study demonstrates the lab’s dramatic progress.
Up to now, scientists would feed chemically synthesized noncanonical amino acids into cells. Having the cell do the work is far more efficient, Xiao said, but that requires the discovery of a new transferase enzyme with tyrosine pockets that could bind sulfate. That lock-and-key combination could then be used as the foundation for a variety of catalysts.
“Now, through this new strategy to modify proteins, we can totally change a protein’s structure and its function,” he said. “For our thrombin inhibitors models, we showed that putting an unnatural building block in the drug can make the drug much more potent.”
It was worth a look to see if nature had beaten them to a useful codon. For that, Xiao enlisted Wolynes, co-director of the Center for Theoretical Biological Physics, whose lab compared genome databases and found sulfotransferase 1C1 in the ibis.
The Xiao lab employed a mutant amber stop codon, a three-nucleotide group of uracil, adenine and guanine, to encode the desired sulfotransferase, resulting in a completely autonomous mammalian cell line capable of biosynthesizing sTyr and incorporating it with great precision into proteins.
“We got lucky,” Xiao said. “Ibis is the only species doing this, which was discovered by a sequence similarity search of genomic information. After that, we asked if they can figure out why this enzyme recognizes tyrosine but our human sulfotransferase cannot.”
The Wolynes team employed AlphaFold2, an artificial intelligence program developed by Alphabet/Google’s DeepMind that predicts proteins structures.
The researchers expect to use the combination of bioinformatics and computationally enhanced screening to produce a library of biosynthesized noncanonical amino acids.